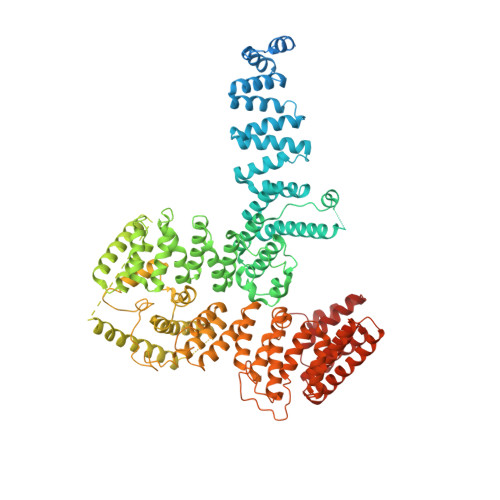
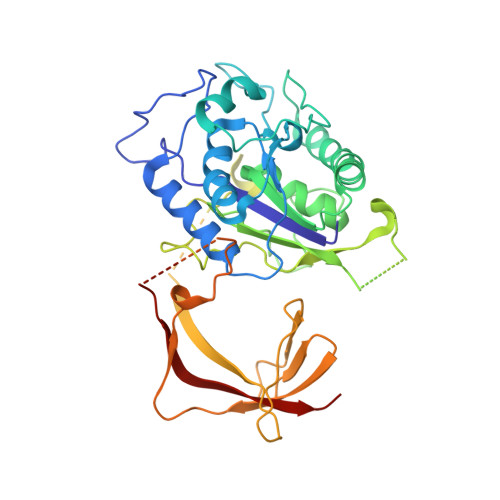
Structure of the TFIIIC subcomplex tau A provides insights into RNA polymerase III pre-initiation complex formation.
Vorlander, M.K., Jungblut, A., Karius, K., Baudin, F., Grotsch, H., Kosinski, J., Muller, C.W.(2020) Nat Commun 11: 4905-4905
- PubMed: 32999288
- DOI: https://doi.org/10.1038/s41467-020-18707-y
- Primary Citation of Related Structures:
6YJ6 - PubMed Abstract:
Transcription factor (TF) IIIC is a conserved eukaryotic six-subunit protein complex with dual function. It serves as a general TF for most RNA polymerase (Pol) III genes by recruiting TFIIIB, but it is also involved in chromatin organization and regulation of Pol II genes through interaction with CTCF and condensin II. Here, we report the structure of the S. cerevisiae TFIIIC subcomplex τA, which contains the most conserved subunits of TFIIIC and is responsible for recruitment of TFIIIB and transcription start site (TSS) selection at Pol III genes. We show that τA binding to its promoter is auto-inhibited by a disordered acidic tail of subunit τ95. We further provide a negative-stain reconstruction of τA bound to the TFIIIB subunits Brf1 and TBP. This shows that a ruler element in τA achieves positioning of TFIIIB upstream of the TSS, and suggests remodeling of the complex during assembly of TFIIIB by TFIIIC.
Organizational Affiliation:
European Molecular Biology Laboratory (EMBL), Structural and Computational Biology Unit, Meyerhofstrasse 1, 69117, Heidelberg, Germany.