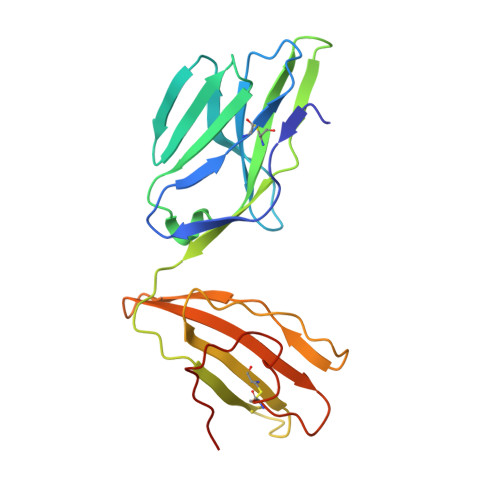
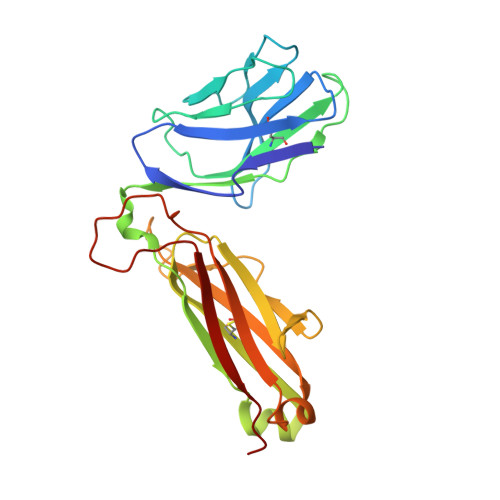
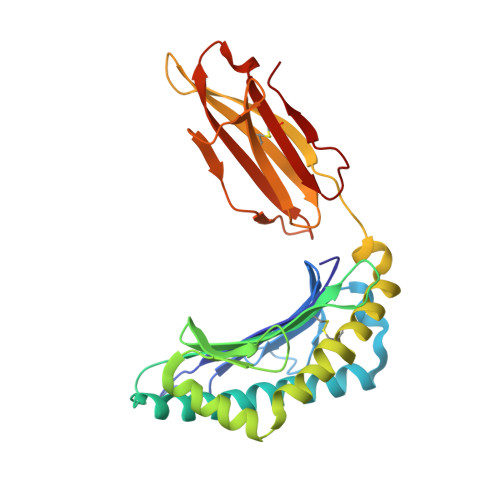
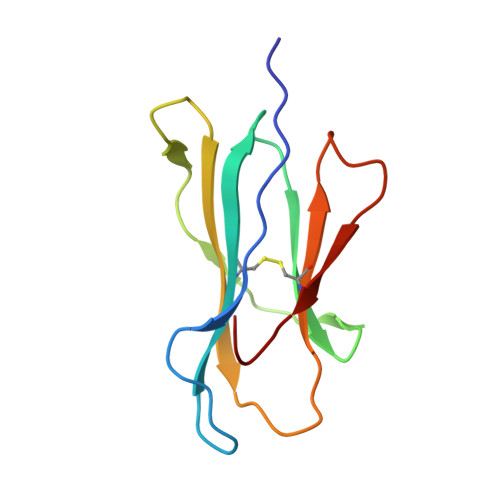
Structurally silent peptide anchor modifications allosterically modulate T cell recognition in a receptor-dependent manner
Smith, A.R., Alonso, J.A., Ayres, C.M., Singh, N.K., Hellman, L.M., Baker, B.M.(2021) Proc Natl Acad Sci U S A 118: e2018125118