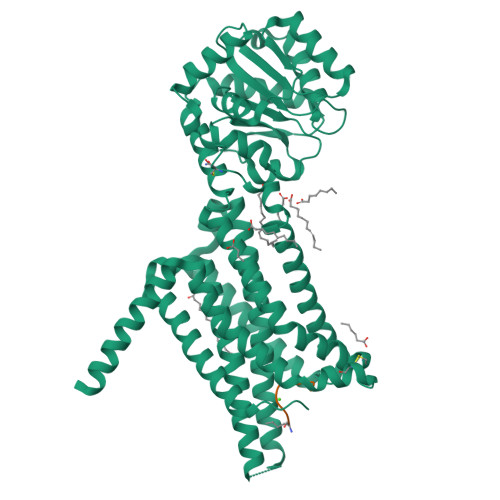
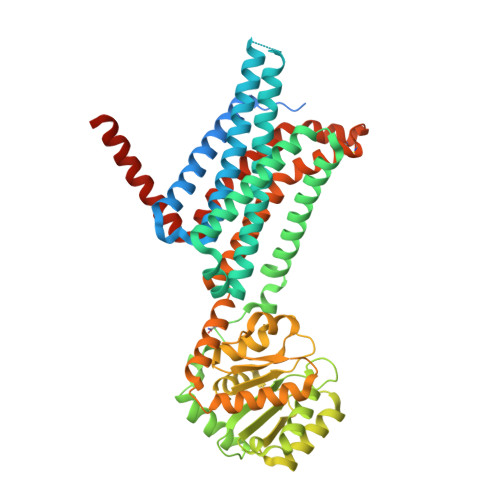
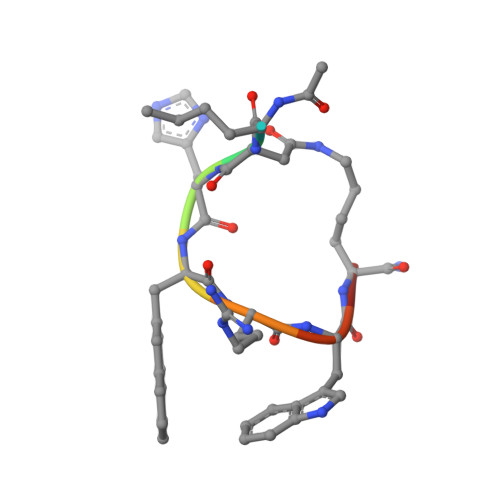
Determination of the melanocortin-4 receptor structure identifies Ca2+as a cofactor for ligand binding.
Yu, J., Gimenez, L.E., Hernandez, C.C., Wu, Y., Wein, A.H., Han, G.W., McClary, K., Mittal, S.R., Burdsall, K., Stauch, B., Wu, L., Stevens, S.N., Peisley, A., Williams, S.Y., Chen, V., Millhauser, G.L., Zhao, S., Cone, R.D., Stevens, R.C.(2020) Science 368: 428-433
- PubMed: 32327598
- DOI: https://doi.org/10.1126/science.aaz8995
- Primary Citation of Related Structures:
6W25 - PubMed Abstract:
The melanocortin-4 receptor (MC4R) is involved in energy homeostasis and is an important drug target for syndromic obesity. We report the structure of the antagonist SHU9119-bound human MC4R at 2.8-angstrom resolution. Ca 2+ is identified as a cofactor that is complexed with residues from both the receptor and peptide ligand. Extracellular Ca 2+ increases the affinity and potency of the endogenous agonist α-melanocyte-stimulating hormone at the MC4R by 37- and 600-fold, respectively. The ability of the MC4R crystallized construct to couple to ion channel Kir7.1, while lacking cyclic adenosine monophosphate stimulation, highlights a heterotrimeric GTP-binding protein (G protein)-independent mechanism for this signaling modality. MC4R is revealed as a structurally divergent G protein-coupled receptor (GPCR), with more similarity to lipidic GPCRs than to the homologous peptidic GPCRs.
Organizational Affiliation:
iHuman Institute, ShanghaiTech University, Pudong, Shanghai 201210, China.