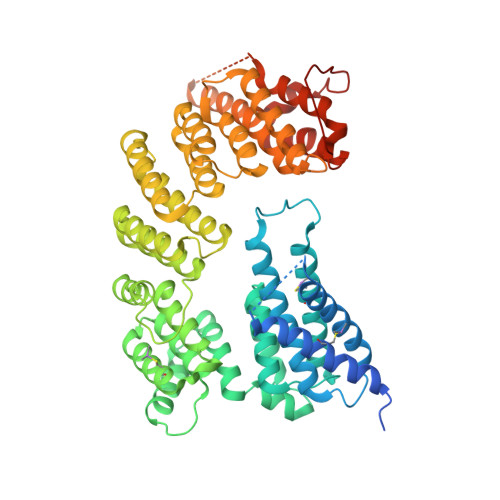
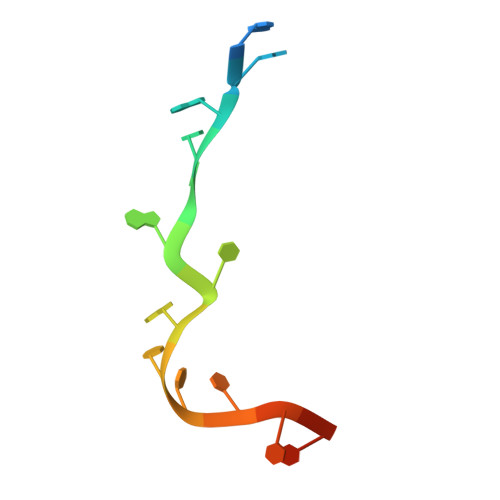
The pentatricopeptide repeat protein Rmd9 recognizes the dodecameric element in the 3'-UTRs of yeast mitochondrial mRNAs.
Hillen, H.S., Markov, D.A., Wojtas, I.D., Hofmann, K.B., Lidschreiber, M., Cowan, A.T., Jones, J.L., Temiakov, D., Cramer, P., Anikin, M.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33876744
- DOI: https://doi.org/10.1073/pnas.2009329118
- Primary Citation of Related Structures:
7A9W, 7A9X - PubMed Abstract:
Stabilization of messenger RNA is an important step in posttranscriptional gene regulation. In the nucleus and cytoplasm of eukaryotic cells it is generally achieved by 5' capping and 3' polyadenylation, whereas additional mechanisms exist in bacteria and organelles. The mitochondrial mRNAs in the yeast Saccharomyces cerevisiae comprise a dodecamer sequence element that confers RNA stability and 3'-end processing via an unknown mechanism. Here, we isolated the protein that binds the dodecamer and identified it as Rmd9, a factor that is known to stabilize yeast mitochondrial RNA. We show that Rmd9 associates with mRNA around dodecamer elements in vivo and that recombinant Rmd9 specifically binds the element in vitro. The crystal structure of Rmd9 bound to its dodecamer target reveals that Rmd9 belongs to the family of pentatricopeptide (PPR) proteins and uses a previously unobserved mode of specific RNA recognition. Rmd9 protects RNA from degradation by the mitochondrial 3'-exoribonuclease complex mtEXO in vitro, indicating that recognition and binding of the dodecamer element by Rmd9 confers stability to yeast mitochondrial mRNAs.
Organizational Affiliation:
Department of Molecular Biology, Max Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany.