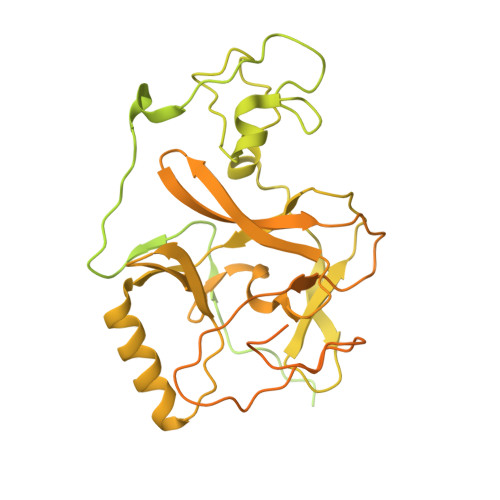
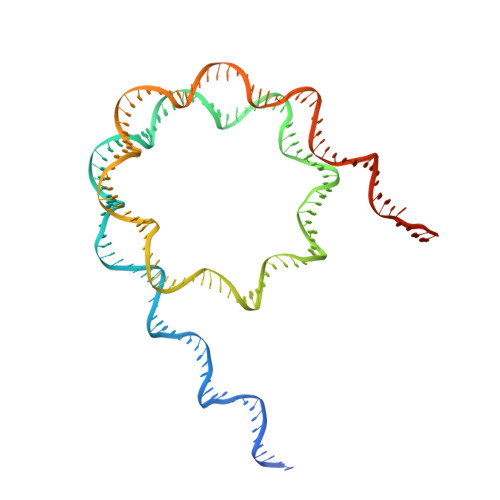
Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Li, W., Tian, W., Yuan, G., Deng, P., Sengupta, D., Cheng, Z., Cao, Y., Ren, J., Qin, Y., Zhou, Y., Jia, Y., Gozani, O., Patel, D.J., Wang, Z.(2021) Nature 590: 498-503
- PubMed: 33361816
- DOI: https://doi.org/10.1038/s41586-020-03069-8
- Primary Citation of Related Structures:
7CRO, 7CRP, 7CRQ, 7CRR - PubMed Abstract:
Histone methyltransferases of the nuclear receptor-binding SET domain protein (NSD) family, including NSD1, NSD2 and NSD3, have crucial roles in chromatin regulation and are implicated in oncogenesis 1,2 . NSD enzymes exhibit an autoinhibitory state that is relieved by binding to nucleosomes, enabling dimethylation of histone H3 at Lys36 (H3K36) 3-7 . However, the molecular basis that underlies this mechanism is largely unknown. Here we solve the cryo-electron microscopy structures of NSD2 and NSD3 bound to mononucleosomes. We find that binding of NSD2 and NSD3 to mononucleosomes causes DNA near the linker region to unwrap, which facilitates insertion of the catalytic core between the histone octamer and the unwrapped segment of DNA. A network of DNA- and histone-specific contacts between NSD2 or NSD3 and the nucleosome precisely defines the position of the enzyme on the nucleosome, explaining the specificity of methylation to H3K36. Intermolecular contacts between NSD proteins and nucleosomes are altered by several recurrent cancer-associated mutations in NSD2 and NSD3. NSDs that contain these mutations are catalytically hyperactive in vitro and in cells, and their ectopic expression promotes the proliferation of cancer cells and the growth of xenograft tumours. Together, our research provides molecular insights into the nucleosome-based recognition and histone-modification mechanisms of NSD2 and NSD3, which could lead to strategies for therapeutic targeting of proteins of the NSD family.
Organizational Affiliation:
Department of Biology, Cryo-EM Centre, Southern University of Science and Technology, Shenzhen, China.