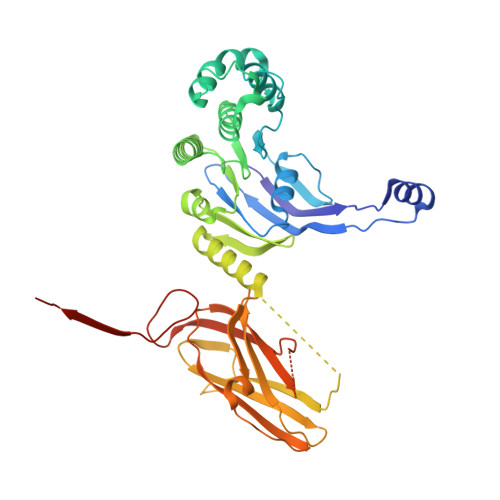
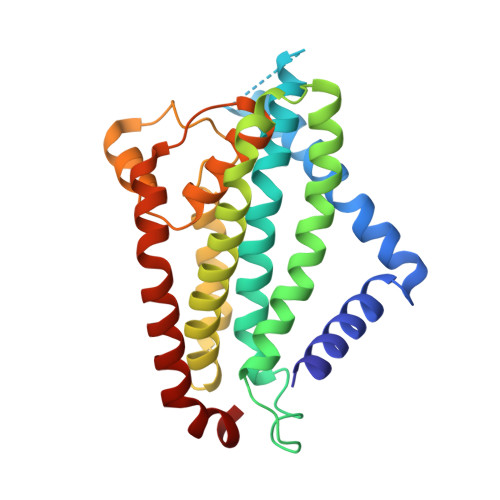
Cryo-EM structure of the full-length WzmWzt ABC transporter required for lipid-linked O antigen transport.
Caffalette, C.A., Zimmer, J.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33443152
- DOI: https://doi.org/10.1073/pnas.2016144118
- Primary Citation of Related Structures:
7K2T - PubMed Abstract:
O antigens are important cell surface polysaccharides in gram-negative bacteria where they extend core lipopolysaccharides in the extracellular leaflet of the outer membrane. O antigen structures are serotype specific and form extended cell surface barriers endowing many pathogens with survival benefits. In the ABC transporter-dependent biosynthesis pathway, O antigens are assembled on the cytosolic side of the inner membrane on a lipid anchor and reoriented to the periplasmic leaflet by the channel-forming WzmWzt ABC transporter for ligation to the core lipopolysaccharides. In many cases, this process depends on the chemical modification of the O antigen's nonreducing terminus, sensed by WzmWzt via a carbohydrate-binding domain (CBD) that extends its nucleotide-binding domain (NBD). Here, we provide the cryo-electron microscopy structure of the full-length WzmWzt transporter from Aquifex aeolicus bound to adenosine triphosphate (ATP) and in a lipid environment, revealing a highly asymmetric transporter organization. The CBDs dimerize and associate with only one NBD. Conserved loops at the CBD dimer interface straddle a conserved peripheral NBD helix. The CBD dimer is oriented perpendicularly to the NBDs and its putative ligand-binding sites face the transporter to likely modulate ATPase activity upon O antigen binding. Further, our structure reveals a closed WzmWzt conformation in which an aromatic belt near the periplasmic channel exit seals the transporter in a resting, ATP-bound state. The sealed transmembrane channel is asymmetric, with one open and one closed cytosolic and periplasmic portal. The structure provides important insights into O antigen recruitment to and translocation by WzmWzt and related ABC transporters.
Organizational Affiliation:
Molecular Physiology and Biological Physics, University of Virginia School of Medicine, Charlottesville, VA 22908.