Full Text |
QUERY: PubMed ID = 23019218 | MyPDB Login | Search API |
| Search Summary | This query matches 27 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameSymmetry TypeSCOP Classification | 1 to 25 of 27 Structures Page 1 of 2 Sort by
From Structure to Function: Insights into the Catalytic Substrate Specificity and Thermostability Displayed by Bacillus subtilis mannanase BCman(2008) J Mol Biol 379: 535-544
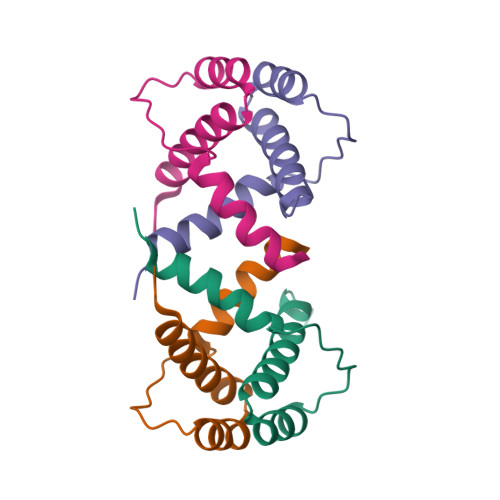
Structure and biochemical studies of the recombination mediator protein RecR in RecFOR pathwayTang, Q., Yan, X.X., Liang, D.C. (2012) Nucleic Acids Res 40: 11115-11125
Structure of recombination mediator protein RecRK21G mutantTang, Q., Yan, X.X., Liang, D.C. (2012) Nucleic Acids Res 40: 11115-11125
Structure of recombination mediator protein RecR16-196 deletion mutantTang, Q., Yan, X.X., Liang, D.C. (2012) Nucleic Acids Res 40: 11115-11125
Crystal structures of N-acyl homoserine lactonase AidHLiang, D.C., Yan, X.X., Gao, A. (2013) Acta Crystallogr D Biol Crystallogr 69: 82-91
Crystal structures of N-acyl homoserine lactonase AidH S102G mutant complexed with N-hexanoyl homoserine lactoneLiang, D.C., Yan, X.X., Gao, A. (2013) Acta Crystallogr D Biol Crystallogr 69: 82-91
Crystal structures of N-acyl homoserine lactonase AidH E219G mutant complexed with N-hexanoyl homoserineLiang, D.C., Yan, X.X., Gao, A. (2013) Acta Crystallogr D Biol Crystallogr 69: 82-91
Crystal structures of N-acyl homoserine lactonase AidH S102G mutantLiang, D.C., Yan, X.X., Gao, A. (2013) Acta Crystallogr D Biol Crystallogr 69: 82-91
Crystal structures of N-acyl homoserine lactonase AidH complexed with N-butanoyl homoserineLiang, D.C., Yan, X.X., Gao, A. (2013) Acta Crystallogr D Biol Crystallogr 69: 82-91
Crystal structures of N-acyl homoserine lactonase AidH E219G mutantLiang, D.C., Yan, X.X., Gao, A. (2013) Acta Crystallogr D Biol Crystallogr 69: 82-91
Crystal Structure of E.coli SbcD at 1.8 A ResolutionLiu, S., Tian, L.F., Yan, X.X., Liang, D.C. (2014) Acta Crystallogr D Biol Crystallogr 70: 299-309
Crystal structure of E.coli SbcD at 2.5 angstrom resolutionLiu, S., Tian, L.F., Yan, X.X., Liang, D.C. (2014) Acta Crystallogr D Biol Crystallogr 70: 299-309
Crystal structure of E.coli SbcD with Mn2+Liu, S., Tian, L.F., Yan, X.X., Liang, D.C. (2014) Acta Crystallogr D Biol Crystallogr 70: 299-309
Structural and functional studies the characterization of Cys4 Zinc-finger motif in the recombination mediator protein RecRTang, Q., Liu, Y.P., Yan, X.X., Liang, D.C. (2014) DNA Repair (Amst) 24: 10-14
Structural and functional studies the characterization of C58G/C70G mutant in Cys4 Zinc-finger motif in the recombination mediator protein RecRTang, Q., Liu, Y.P., Yan, X.X., Liang, D.C. (2014) DNA Repair (Amst) 24: 10-14
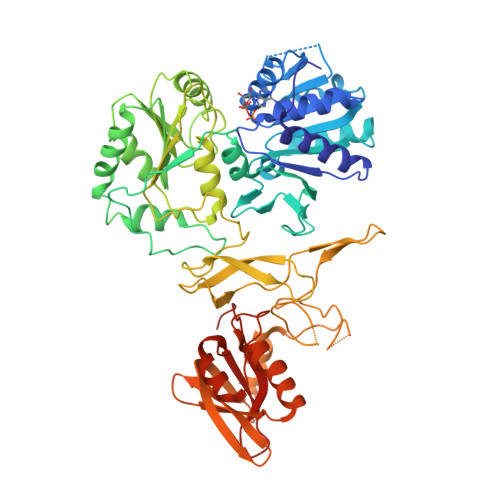
Structural basis underlying complex assembly andconformational transition of the type I R-M systemLiu, Y.P., Tang, Q., Zhang, J.Z., Tian, L.F., Gao, P., Yan, X.X. (2017) Proc Natl Acad Sci U S A 114: 11151-11156
Crystal structure of a disease-related gene, hCDC73(1-111)Sun, W., Kuang, X.L., Liu, Y.P., Tian, L.F., Yan, X.X., Xu, W.Q. (2017) Sci Rep 7: 15638-15638
Crystal structure of a disease-related gene, hCDC73(1-100)Sun, W., Kuang, X.L., Liu, Y.P., Tian, L.F., Yan, X.X., Xu, W.Q. (2017) Sci Rep 7: 15638-15638
Crystal structure of methyladenine demethylaseTian, L.F., Tang, Q., Chen, Z.Z., Yan, X.X. (2020) Cell Res 30: 272-275
Crystal structure of methyladenine demethylaseTian, L.F., Tang, Q., Chen, Z.Z., Yan, X.X. (2020) Cell Res 30: 272-275
Crystal structure of a single strand DNA binding protein(2019) Protein Sci 28: 788-793
crystal structure of a DNA repair protein(2019) Proteins 87: 791-795
Crystal structure of DltDYan, X.X., Zeng, Q., Tian, L.F. (2022) PROG BIOCHEM BIOPHYS 48: 1052-1062
Crystal structure of RadD- ADP complex(2023) Int J Mol Sci 24:
Crystal structure of RadD/ATP analogue complex(2023) Int J Mol Sci 24:
1 to 25 of 27 Structures Page 1 of 2 Sort by |