Catalytic mechanism of heparinase II investigated by site-directed mutagenesis and the crystal structure with its substrate.
Shaya, D., Zhao, W., Garron, M.L., Xiao, Z., Cui, Q., Zhang, Z., Sulea, T., Linhardt, R.J., Cygler, M.(2010) J Biol Chem 285: 20051-20061
- PubMed: 20404324
- DOI: https://doi.org/10.1074/jbc.M110.101071
- Primary Citation of Related Structures:
3E7J, 3E80 - PubMed Abstract:
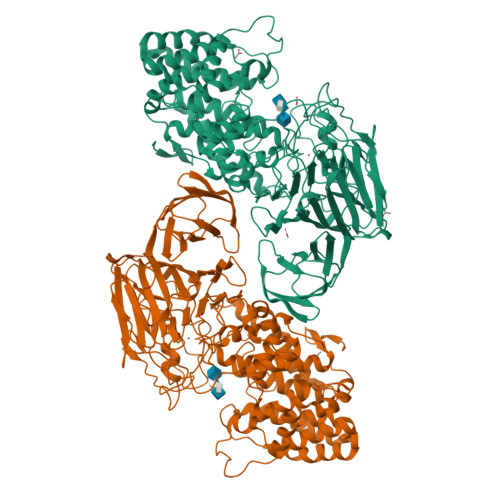
Heparinase II (HepII) is an 85-kDa dimeric enzyme that depolymerizes both heparin and heparan sulfate glycosaminoglycans through a beta-elimination mechanism. Recently, we determined the crystal structure of HepII from Pedobacter heparinus (previously known as Flavobacterium heparinum) in complex with a heparin disaccharide product, and identified the location of its active site. Here we present the structure of HepII complexed with a heparan sulfate disaccharide product, proving that the same binding/active site is responsible for the degradation of both uronic acid epimers containing substrates. The key enzymatic step involves removal of a proton from the C5 carbon (a chiral center) of the uronic acid, posing a topological challenge to abstract the proton from either side of the ring in a single active site. We have identified three potential active site residues equidistant from C5 and located on both sides of the uronate product and determined their role in catalysis using a set of defined tetrasaccharide substrates. HepII H202A/Y257A mutant lost activity for both substrates and we determined its crystal structure complexed with a heparan sulfate-derived tetrasaccharide. Based on kinetic characterization of various mutants and the structure of the enzyme-substrate complex we propose residues participating in catalysis and their specific roles.
Organizational Affiliation:
Department of Biochemistry, McGill University, Montréal, Québec H3G 1Y6, Canada.