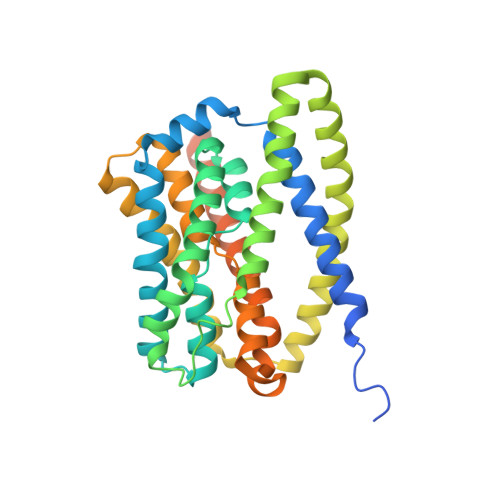
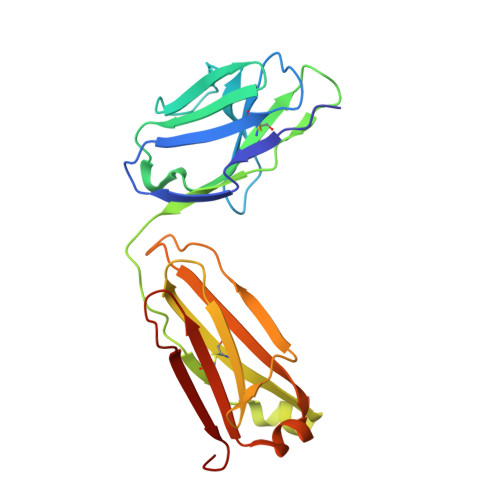
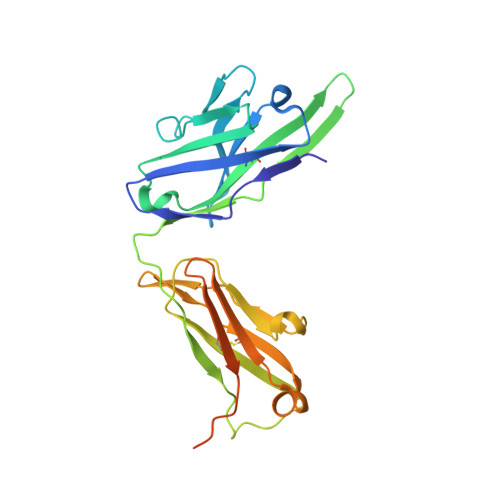
Structural insights into the HBV receptor and bile acid transporter NTCP.
Park, J.H., Iwamoto, M., Yun, J.H., Uchikubo-Kamo, T., Son, D., Jin, Z., Yoshida, H., Ohki, M., Ishimoto, N., Mizutani, K., Oshima, M., Muramatsu, M., Wakita, T., Shirouzu, M., Liu, K., Uemura, T., Nomura, N., Iwata, S., Watashi, K., Tame, J.R.H., Nishizawa, T., Lee, W., Park, S.Y.(2022) Nature 606: 1027-1031
- PubMed: 35580630
- DOI: https://doi.org/10.1038/s41586-022-04857-0
- Primary Citation of Related Structures:
7FCI - PubMed Abstract:
Around 250 million people are infected with hepatitis B virus (HBV) worldwide 1 , and 15 million may also carry the satellite virus hepatitis D virus (HDV), which confers even greater risk of severe liver disease 2 . The HBV receptor has been identified as sodium taurocholate co-transporting polypeptide (NTCP), which interacts directly with the first 48 amino acid residues of the N-myristoylated N-terminal preS1 domain of the viral large protein 3 . Despite the pressing need for therapeutic agents to counter HBV, the structure of NTCP remains unsolved. This 349-residue protein is closely related to human apical sodium-dependent bile acid transporter (ASBT), another member of the solute carrier family SLC10. Crystal structures have been reported of similar bile acid transporters from bacteria 4,5 , and these models are believed to resemble closely both NTCP and ASBT. Here we have used cryo-electron microscopy to solve the structure of NTCP bound to an antibody, clearly showing that the transporter has no equivalent of the first transmembrane helix found in other SLC10 proteins, and that the N terminus is exposed on the extracellular face. Comparison of our structure with those of related proteins indicates a common mechanism of bile acid transport, but the NTCP structure displays an additional pocket formed by residues that are known to interact with preS1, presenting new opportunities for structure-based drug design.
Organizational Affiliation:
Drug Design Laboratory, Graduate School of Medical Life Science, Yokohama City University, Yokohama, Japan.