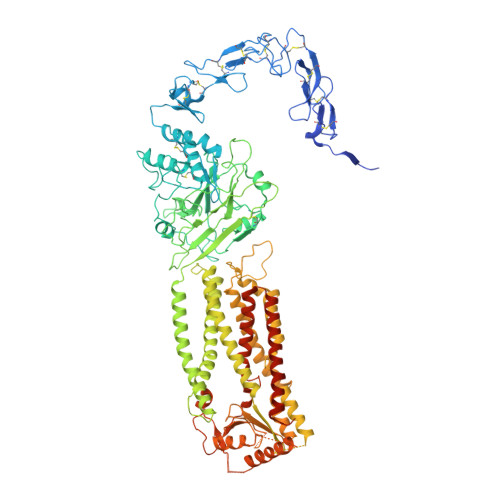
Structure of the human Meckel-Gruber protein Meckelin.
Liu, D., Qian, D., Shen, H., Gong, D.(2021) Sci Adv 7: eabj9748-eabj9748
- PubMed: 34731008
- DOI: https://doi.org/10.1126/sciadv.abj9748
- Primary Citation of Related Structures:
7FH1 - PubMed Abstract:
Mutations in the Meckelin gene account for most cases of the Meckel-Gruber syndrome, the most severe ciliopathy with a 100% mortality rate. Here, we report a 3.3-Å cryo–electron microscopy structure of human Meckelin (also known as TMEM67 and MKS3). The structure reveals a unique protein fold consisting of an unusual cysteine-rich domain that folds as an arch bridge stabilized by 11 pairs of disulfide bonds, a previously uncharacterized domain named β sheet–rich domain, a previously unidentified seven-transmembrane fold wherein TM4 to TM6 are broken near the cytoplasmic surface of the membrane, and a coiled-coil domain placed below the transmembrane domain. Meckelin forms a stable homodimer with an extensive dimer interface. Our structure establishes a framework for dissecting the function and disease mechanisms of Meckelin.
Organizational Affiliation:
Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang 310024, China.