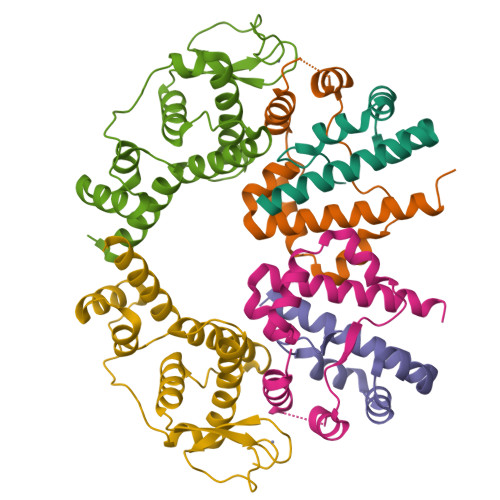
Hexameric structure of the flagellar master regulator FlhDC from Cupriavidus necator and its interaction with flagellar promoter DNA.
Cho, S.Y., Oh, H.B., Yoon, S.I.(2023) Biochem Biophys Res Commun 672: 97-102
- PubMed: 37343320
- DOI: https://doi.org/10.1016/j.bbrc.2023.06.033
- Primary Citation of Related Structures:
8J56 - PubMed Abstract:
Bacterial flagella are assembled with ∼30 different proteins in a defined order via diverse regulatory systems. In gram-negative bacteria from the Gammaproteobacteria and Betaproteobacteria classes, the transcription of flagellar genes is strictly controlled by the master regulator FlhDC. In Gammaproteobacteria species, the FlhDC complex has been shown to activate flagellar expression by directly interacting with the promoter region in flagellar genes. To obtain the DNA-binding mechanism of FlhDC and determine the conserved and distinct structural features of Betaproteobacteria and Gammaproteobacteria FlhDCs that are necessary for their functions, we determined the crystal structure of Betaproteobacteria Cupriavidus necator FlhDC (cnFlhDC) and biochemically analyzed its DNA-binding capacity. cnFlhDC specifically recognized the promoter DNA of the class II flagellar genes flgB and flhB. cnFlhDC adopts a ring-like heterohexameric structure (cnFlhD 4 C 2 ) and harbors two Zn-Cys clusters, as observed for Gammaproteobacteria Escherichia coli FlhDC (ecFlhDC). The cnFlhDC structure exhibits positively charged surfaces across two FlhDC subunits as a putative DNA-binding site. Noticeably, the positive patch of cnFlhDC is continuous, in contrast to the separated patches of ecFlhDC. Moreover, the ternary intersection of cnFlhD 4 C 2 behind the Zn-Cys cluster forms a unique protruding neutral structure, which is replaced with a charged cavity in the ecFlhDC structure.
Organizational Affiliation:
Division of Biomedical Convergence, College of Biomedical Science, Kangwon National University, Chuncheon, 24341, Republic of Korea.