Crystal structure of a novel PU plastic degradation enzyme with ligand from Thermaerobacter marianensis
Li, Z.S., Wang, H., Zheng, Z.R., Cong, L., Chen, Y.Y., Han, X., Wei, R., Uwe, B., liu, W.D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Carboxylic ester hydrolase | 527 | Thermaerobacter marianensis DSM 12885 | Mutation(s): 1 Gene Names: Tmar_0630 EC: 3.1.1 | 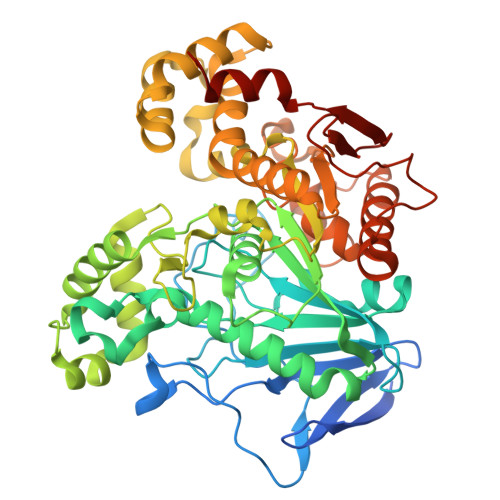 | |
UniProt | |||||
Find proteins for E6SHQ4 (Thermaerobacter marianensis (strain ATCC 700841 / DSM 12885 / JCM 10246 / 7p75a)) Explore E6SHQ4 Go to UniProtKB: E6SHQ4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | E6SHQ4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A1D5F (Subject of Investigation/LOI) Query on A1D5F | F [auth C] | 4-oxidanylbutyl ~{N}-[4-[(4-aminophenyl)methyl]phenyl]carbamate C18 H22 N2 O3 UWOCPQGJYMGPEI-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | E [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 81.566 | α = 90 |
| b = 81.566 | β = 90 |
| c = 667.529 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | -- |