Suppressing Tymovirus replication in plants using a variant of ubiquitin.
De Silva, A., Kim, K., Weiland, J., Hwang, J., Chung, J., Pereira, H.S., Patel, T.R., Teyra, J., Patel, A., Mira, M.M., Khajehpour, M., Bolton, M., Stasolla, C., Sidhu, S.S., Mark, B.L.(2025) PLoS Pathog 21: e1012899-e1012899
- PubMed: 39869641
- DOI: https://doi.org/10.1371/journal.ppat.1012899
- Primary Citation of Related Structures:
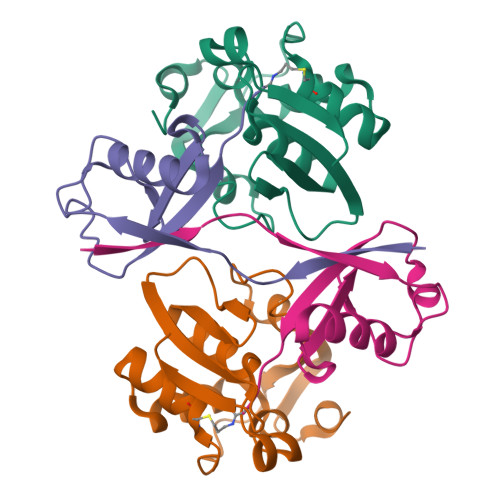
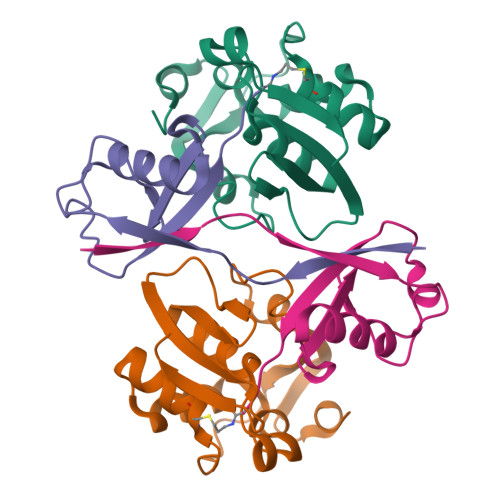
9CSF, 9CSH - PubMed Abstract:
RNA viruses have evolved numerous strategies to overcome host resistance and immunity, including the use of multifunctional proteases that not only cleave viral polyproteins during virus replication but also deubiquitinate cellular proteins to suppress ubiquitin (Ub)-mediated antiviral mechanisms. Here, we report an approach to attenuate the infection of Arabidopsis thaliana by Turnip Yellow Mosaic Virus (TYMV) by suppressing the polyprotein cleavage and deubiquitination activities of the TYMV protease (PRO). Performing selections using a library of phage-displayed Ub variants (UbVs) for binding to recombinant PRO yielded several UbVs that bound the viral protease with nanomolar affinities and blocked its function. The strongest binding UbV (UbV3) candidate had a EC50 of 0.3 nM and inhibited both polyprotein cleavage and DUB activity of PRO in vitro. X-ray crystal structures of UbV3 alone and in complex with PRO reveal that the inhibitor exists as a dimer that binds two copies of PRO. Consistent with our biochemical and structural findings, transgenic expression of UbV3 in the cytosol of A. thaliana suppressed TYMV replication in planta, with the reduction in viral load being correlated to UbV3 expression level. Our results demonstrate the potential of using UbVs to protect plants from tymovirus infection, a family of viruses that contain numerous members of significant agricultural concern, as well as other plant viruses that express functionally related proteases with deubiquitinating activity.
Organizational Affiliation:
Department of Microbiology, Faculty of Science, University of Manitoba, Winnipeg, Manitoba, Canada.