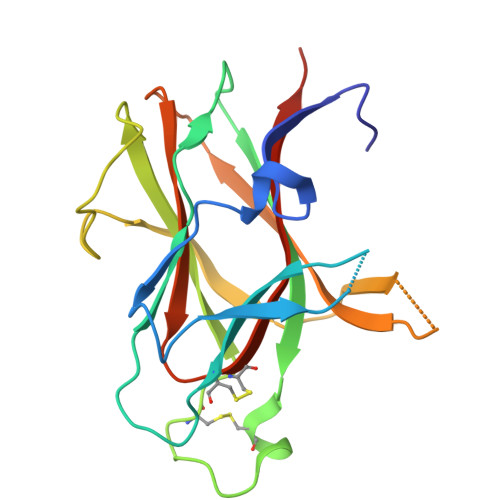
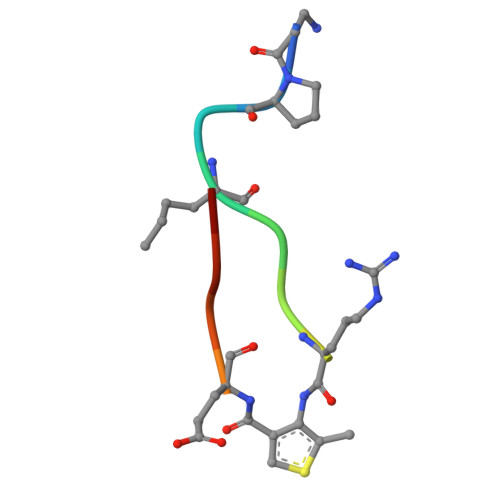
Constrained beta-Hairpins Targeting the EphA4 Ligand Binding Domain.
Prentiss, A.M., Baggio, C., Pagett, J., Kulinich, A.O., Ethell, I.M., Muzzarelli, K., Assar, Z., Pellecchia, M.(2024) J Med Chem 67: 22245-22253
- PubMed: 39656022
- DOI: https://doi.org/10.1021/acs.jmedchem.4c02286
- Primary Citation of Related Structures:
9CY8 - PubMed Abstract:
The activity of the receptor tyrosine kinase EphA4 has been implicated in several pathologies including oncology (gastric and pancreatic cancers) and neurodegenerative diseases (amyotrophic lateral sclerosis and Alzheimer's disease). However, advances in validating EphA4 as a possible drug target have been limited by the lack of suitable pharmacological inhibitors. Recently, we reported on the design of potent EphA4 agonistic agents targeting its ligand binding domain (LBD). Based on previous studies with a phage display cyclic peptide inhibitor, we designed a β-hairpin mimetic with high affinity for EphA4-LBD. These agents hold great promise for further validation and development of EphA4-based therapeutics. Moreover, our studies introduce a possible strategy for the design of constrained β-hairpin peptides.
Organizational Affiliation:
Division of Biomedical Sciences, School of Medicine, University of California Riverside, 900 University Avenue, Riverside, California 92521, United States.