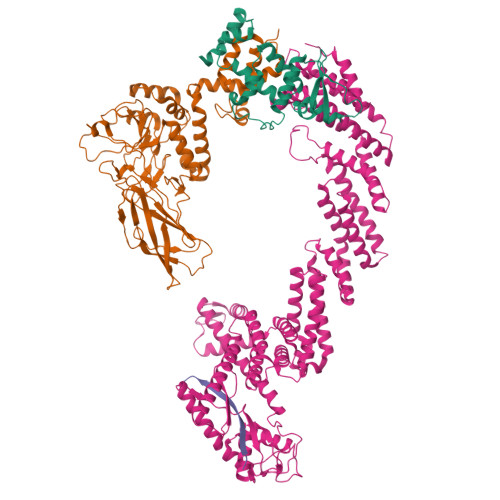
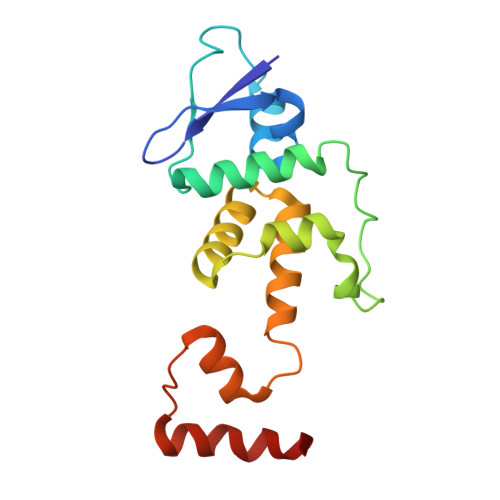
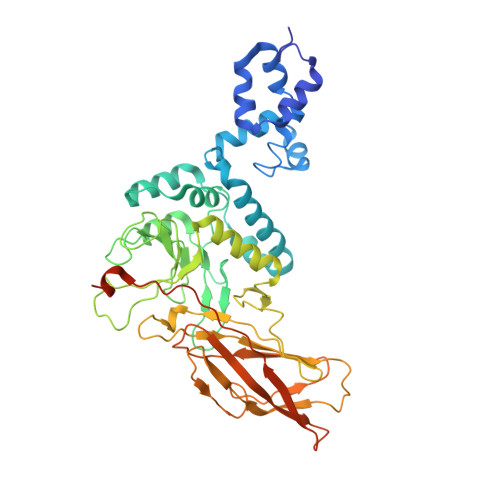
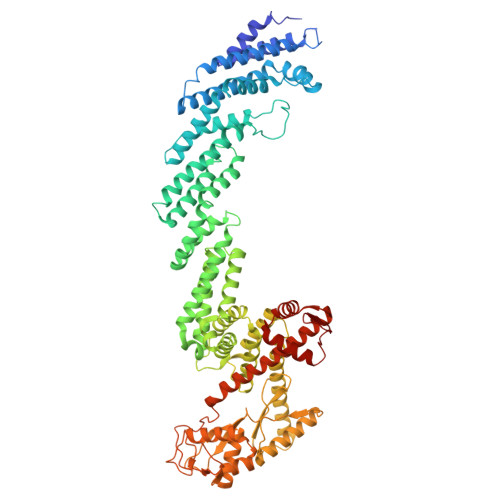
Structural Insight Into the SKP1-CUL1-FBXO3-RBX1 Complex.
Wei, J., Xu, C.(2025) Proteins
- PubMed: 39921442
- DOI: https://doi.org/10.1002/prot.26809
- Primary Citation of Related Structures:
9KBD, 9KBF - PubMed Abstract:
The cryo-EM structure of human SCF FBXO3 , which consists of CUL1, RBX1, SKP1 and FBXO3 was solved at a nominal resolution of 3.70 Å. Although a previous study reported the crystal structure of the FBXO3 ApaG domain, how FBXO3 is incorporated into the SCF complex remains elusive. In the cryo-EM structure of SCF FBXO3 , the F-box domain of FBXO3 primarily associates with SKP1 via extensive hydrophobic interactions and interacts with the N-terminal region of CUL1 via hydrophobic interactions. The weak cryo-EM map of the RBX1 globular region is close to the FBXO3 ApaG domain, suggesting that unmodified SCF FBXO3 exhibits a closed conformation and that CUL1 neddylation is likely required to achieve high E3 activity. The structural study provides insight into the assembly of SCF FBXO3 and its activation mediated by CUL1 neddylation.
Organizational Affiliation:
MOE Key Laboratory for Membraneless Organelles and Cellular Dynamics, Center for Advanced Interdisciplinary Science and Biomedicine of IHM, Hefei National Laboratory for Physical Sciences at the Microscale, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China.