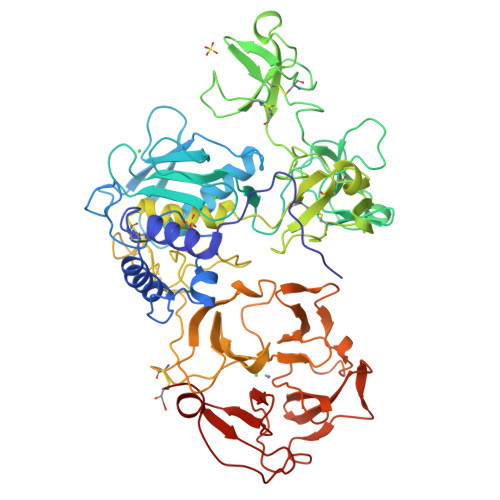
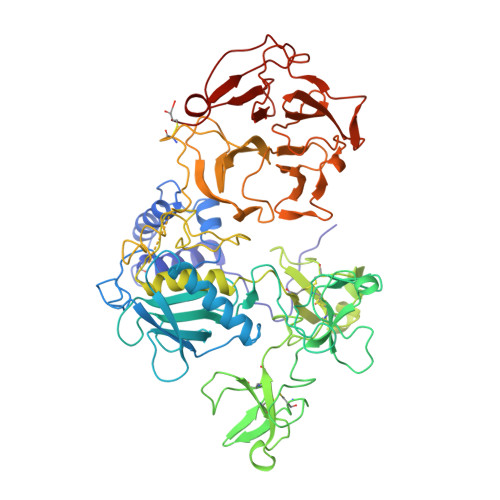
Structure of human pro-matrix metalloproteinase-2: activation mechanism revealed.
Morgunova, E., Tuuttila, A., Bergmann, U., Isupov, M., Lindqvist, Y., Schneider, G., Tryggvason, K.(1999) Science 284: 1667-1670
- PubMed: 10356396
- DOI: https://doi.org/10.1126/science.284.5420.1667
- Primary Citation of Related Structures:
1CK7 - PubMed Abstract:
Matrix metalloproteinases (MMPs) catalyze extracellular matrix degradation. Control of their activity is a promising target for therapy of diseases characterized by abnormal connective tissue turnover. MMPs are expressed as latent proenzymes that are activated by proteolytic cleavage that triggers a conformational change in the propeptide (cysteine switch). The structure of proMMP-2 reveals how the propeptide shields the catalytic cleft and that the cysteine switch may operate through cleavage of loops essential for propeptide stability.
Organizational Affiliation:
Division of Matrix Biology, Department of Medical Biochemistry and Biophysics, Karolinska Institute, Stockholm, Sweden.