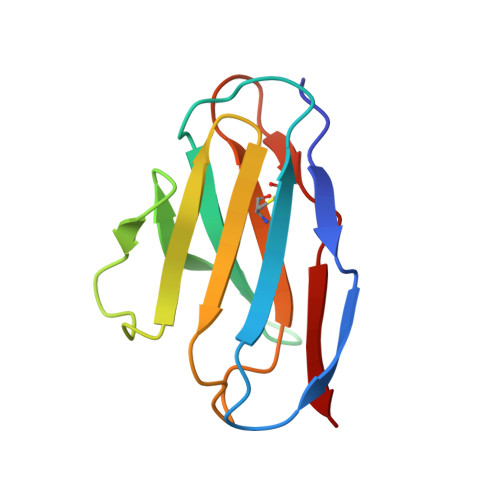
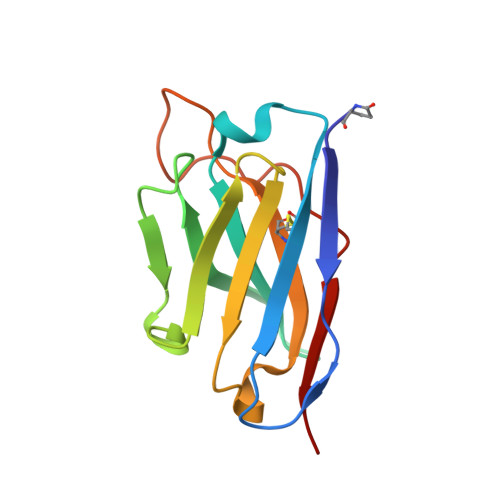
An unusual human IgM antibody with a protruding HCDR3 and high avidity for its peptide ligands.
Ramsland, P.A., Shan, L., Moomaw, C.R., Slaughter, C.A., Fan, Z., Guddat, L.W., Edmundson, A.B.(2000) Mol Immunol 37: 295-310
- PubMed: 11000403
- DOI: https://doi.org/10.1016/s0161-5890(00)00049-3
- Primary Citation of Related Structures:
1DQL - PubMed Abstract:
The crystal structure of the Fv molecule from a human monoclonal IgM cryoglobulin (Mez) was determined at 2.6 A resolution. Amino acid sequences of framework regions (FR) of the Mez light (L) and heavy (H) chain variable domains (VL and VH) are highly similar to their counterparts in another human Fv (Pot) previously subjected to X-ray analysis in our laboratory. As expected, the three-dimensional (3-D) structures of FR are quite similar in the two proteins, as are four of the six complementarity-determining regions (CDRs): CDRs 1 and 2 for both L and H chains. Absence of Pro 95L from the LCDR3 loop in Mez VL (relative to Pot LCDR3) results in compression of this loop and creates more space in the VL-VH interface. In the two IgMs, HCDR3 conformations differ significantly from all previously defined conformations for these loops. Pot has a 12-residue HCDR3 that collapses to fill all available space in the VL-VH domain interface, resulting in the formation of a relatively flat platform for antigen binding. In Mez, the HCDR3 is two residues longer and is comprehensively different. A semi-rigid ascending segment dominated by a Pro-Pro-Tyr sequence protrudes out into solvent. The descending portion has the sequence Gly-Trp-Gly-Gly-Gly, which promotes high local flexibility. This segment folds across the VL-VH domain interface to interact with residues in LCDR3. These features partition the Mez active site into two compartments, a large cavity between VL and VH and a smaller cavity lined entirely by constituents of the three heavy chain CDRs. Such an unusual topographical feature indicates why the Mez IgM does not bind to the Fc portion of intact human IgG antibodies in immunoassays yet interacts with high avidity with many Fc-derived octapeptides. The cavities are expected to be the repositories for the Fc-derived peptides, while the semi-rigid protrusion of the Mez HCDR3 prevents the close approach of another macromolecule (e.g. intact IgG) to the active site.
Organizational Affiliation:
Crystallography Program, Oklahoma Medical Research Foundation, 825 NE 13th Street, Oklahoma City, OK 73104, USA.