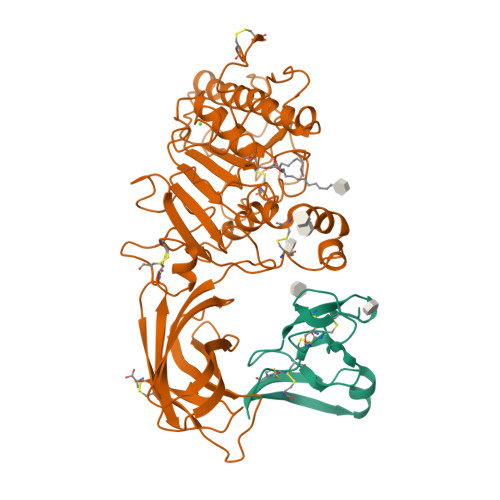
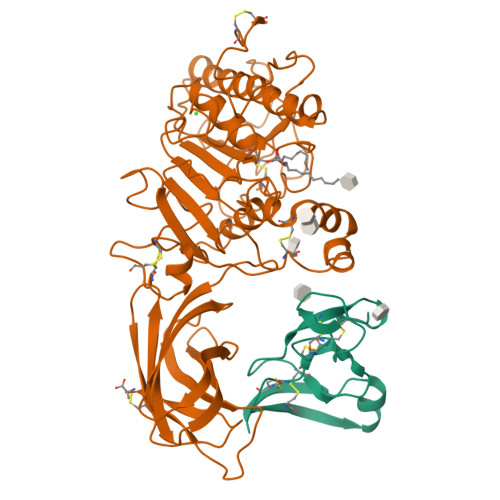
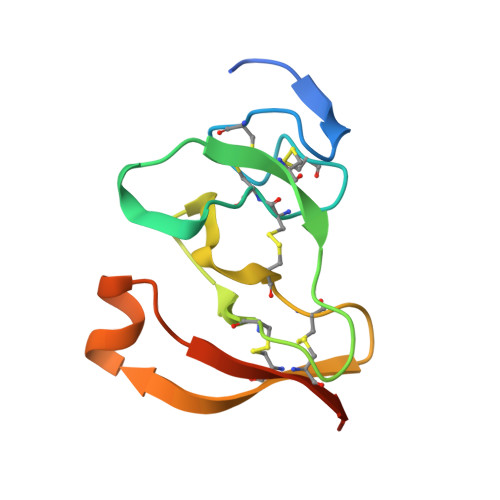
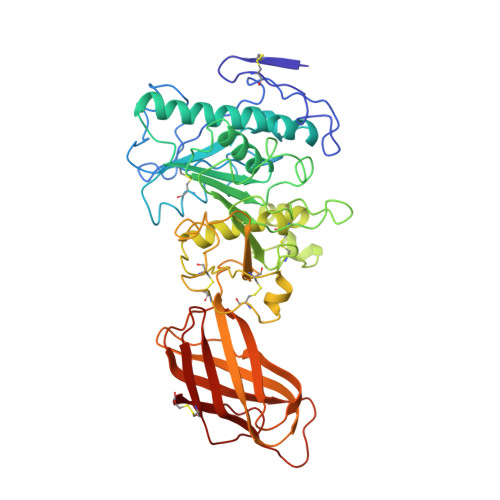
The 2.46 A resolution structure of the pancreatic lipase-colipase complex inhibited by a C11 alkyl phosphonate.
Egloff, M.P., Marguet, F., Buono, G., Verger, R., Cambillau, C., van Tilbeurgh, H.(1995) Biochemistry 34: 2751-2762
- PubMed: 7893686
- DOI: https://doi.org/10.1021/bi00009a003
- Primary Citation of Related Structures:
1LPB - PubMed Abstract:
Pancreatic lipase belongs to the serine esterase family and can therefore be inhibited by classical serine reagents such as diisopropyl fluoride or E600. In an attempt to further characterize the active site and catalytic mechanism, we synthesized a C11 alkyl phosphonate compound. This compound is an effective inhibitor of pancreatic lipase. The crystal structure of the pancreatic lipase-colipase complex inhibited by this compound was determined at a resolution of 2.46 A and refined to a final R-factor of 18.3%. As was observed in the case of the structure of the ternary pancreatic lipase-colipase-phospholipid complex, the binding of the ligand induces rearrangements of two surface loops in comparison with the closed structure of the enzyme (van Tilbeurgh et al., 1993b). The inhibitor, which could be clearly observed in the active site, was covalently bound to the active site serine Ser152. A racemic mixture of the inhibitor was used in the crystallization, and there exists evidence that both enantiomers are bound at the active site. The C11 alkyl chain of the first enantiomer fits into a hydrophobic groove and is though to thus mimic the interaction between the leaving fatty acid of a triglyceride substrate and the protein. The alkyl chain of the second enantiomer also has an elongated conformation and interacts with hydrophobic patches on the surface of the open amphipathic lid. This may indicate the location of a second alkyl chain of a triglyceride substrate. Some of the detergent molecules, needed for the crystallization, were also observed in the crystal. Some of them were located at the entrance of the active site, bound to the hydrophobic part of the lid. On the basis of this crystallographic study, a hypothesis about the binding mode of real substrates and the organization of the active site is proposed.
Organizational Affiliation:
Laboratoire de Cristallisation et Cristallographie des Macromolécules Biologiques, URA 1296-CNRS, Faculté de Médecine Nord, Marseille, France.