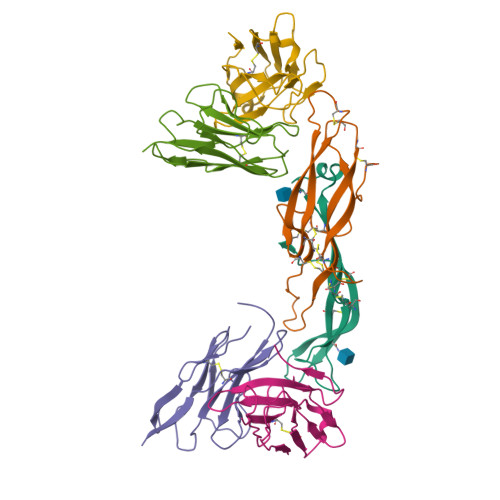
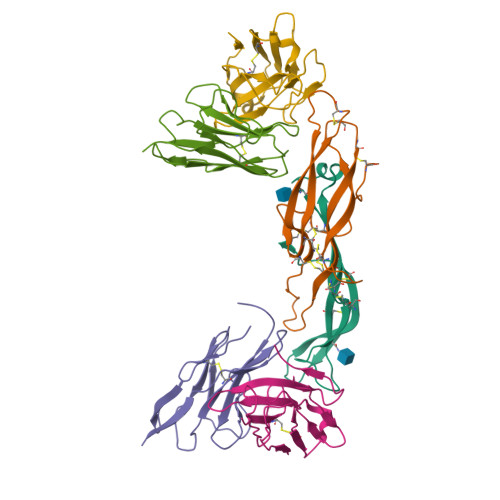
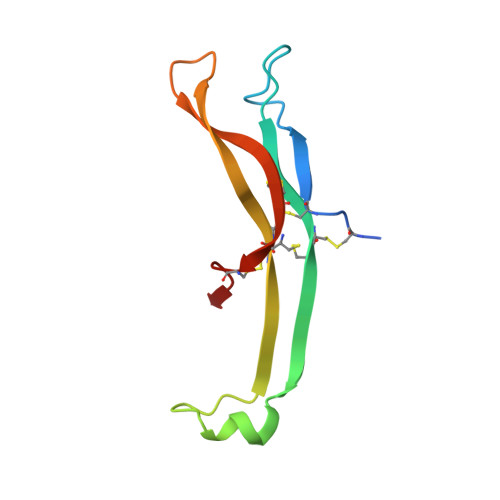
Crystal structure of a ternary complex between human chorionic gonadotropin (hCG) and two Fv fragments specific for the alpha and beta-subunits.
Tegoni, M., Spinelli, S., Verhoeyen, M., Davis, P., Cambillau, C.(1999) J Mol Biology 289: 1375-1385
- PubMed: 10373373
- DOI: https://doi.org/10.1006/jmbi.1999.2845
- Primary Citation of Related Structures:
1QFW - PubMed Abstract:
Human chorionic gonadotropin (hCG), is a placental hormone which exerts its major effect by stimulating progesterone production, crucially sustaining the early weeks of pregnancy. Detection of hCG with specific monoclonal antibodies (mAbs) has become the chosen means for pregnancy diagnosis. We have used antibody Fv fragments derived from two high-affinity mAbs, one against the alpha and the other against the beta-hCG subunit to enable the crystallisation of intact or desialylated hCG. Crystals of a ternary complex composed of Fv anti-alpha/hCG/Fv anti-beta were found to diffract to 3.5 A resolution, and the structure was solved by molecular replacement. In the crystal, the two Fvs keep hCG as in a molecular cage, providing good protein-protein contacts and leaving enough space for the saccharides to be accommodated in the cell solvent. The two Fvs were found not to interact directly through their complementary-determining regions with the hCG saccharides, but only with the protein. The hCG structure in the ternary complex was very close to that of the HF partially deglycosylated hormone, thus indicating that neither the saccharides nor the Fvs had any substantial influence on hormone structure.
Organizational Affiliation:
Architecture et Fonction des Macromolécules Biologiques UPR 9039, CNRS, IFR1, 31 Chemin Joseph Aiguier, Marseille Cedex 20, 13402, France.