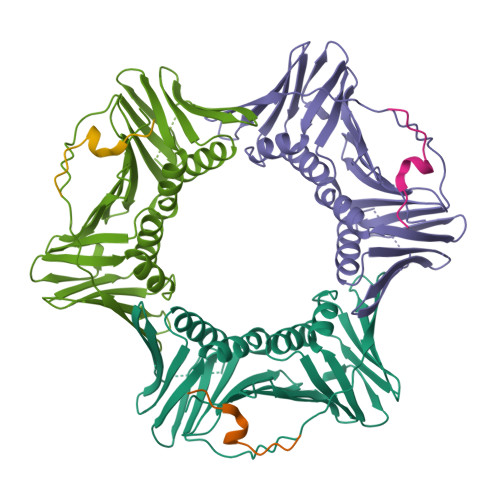
Structural and Thermodynamic Analysis of Human PCNA with Peptides Derived from DNA Polymerase-delta p66 Subunit and Flap Endonuclease-1.
Bruning, J.B., Shamoo, Y.(2004) Structure 12: 2209-2219
- PubMed: 15576034
- DOI: https://doi.org/10.1016/j.str.2004.09.018
- Primary Citation of Related Structures:
1U76, 1U7B - PubMed Abstract:
Human Proliferating Cellular Nuclear Antigen (hPCNA), a member of the sliding clamp family of proteins, makes specific protein-protein interactions with DNA replication and repair proteins through a small peptide motif termed the PCNA-interacting protein, or PIP-box. We solved the structure of hPCNA bound to PIP-box-containing peptides from the p66 subunit of the human replicative DNA polymerase-delta (452-466) at 2.6 A and of the flap endonuclease (FEN1) (331-350) at 1.85 A resolution. Both structures demonstrate that the pol-delta p66 and FEN1 peptides interact with hPCNA at the same site shown to bind the cdk-inhibitor p21(CIP1). Binding studies indicate that peptides from the p66 subunit of the pol-delta holoenzyme and FEN1 bind hPCNA from 189- to 725-fold less tightly than those of p21. Thus, the PIP-box and flanking regions provide a small docking peptide whose affinities can be readily adjusted in accord with biological necessity to mediate the binding of DNA replication and repair proteins to hPCNA.
Organizational Affiliation:
Department of Biochemistry and Cell Biology, Rice University, 6100 South Main Street, MS140, Houston, TX 77005, USA.