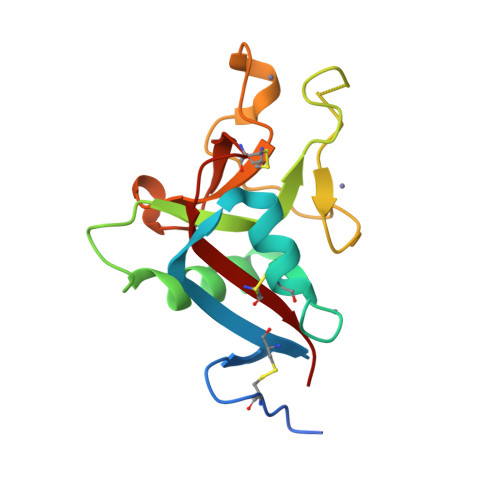
Crystallization and preliminary crystallographic study of HIP/PAP, a human C-lectin overexpressed in primary liver cancers.
Abergel, C., Chenivesse, S., Stinnakre, M.G., Guasco, S., Brechot, C., Claverie, J.M., Devinoy, E., Christa, L.(1999) Acta Crystallogr D Biol Crystallogr 55: 1487-1489
- PubMed: 10417404
- DOI: https://doi.org/10.1107/s0907444999007969
- Primary Citation of Related Structures:
1UV0 - PubMed Abstract:
Human HIP/PAP is an adhesion protein expressed in normal pancreatic and Paneth cells and overexpressed in hepatocellular carcinoma. HIP/PAP was crystallized using the Hampton Research Crystal Screen and SAmBA software to define the optimal crystallization protocol. The crystals belong to the orthorhombic space group P2(1)2(1)2(1), with unit-cell parameters a = 30.73, b = 49.35, c = 92.15 A and one molecule in the asymmetric unit. Flash-frozen crystals diffract to 1. 78 A resolution using synchrotron radiation. A molecular-replacement solution was obtained using the human Reg/lithostathine structure and the AMoRe software.
Organizational Affiliation:
Information Génétique et Structurale, CNRS UMR 1889, Institut de Biologie Structurale et Microbiologie, 31 Chemin Joseph Aiguier, 13402 Marseille CEDEX 20, France. chantal@igs.cnrs-mrs.fr