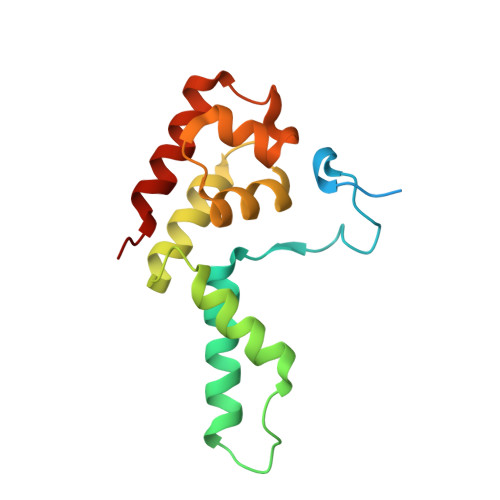
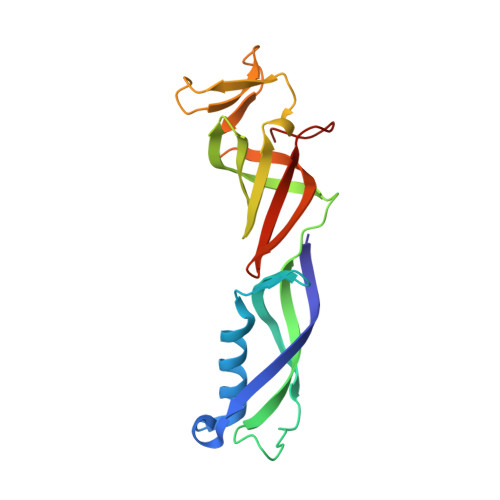
Crystal Structure and RNA Binding of the Rpb4/Rpb7 Subunits of Human RNA Polymerase II.
Meka, H., Werner, F., Cordell, S.C., Onesti, S., Brick, P.(2005) Nucleic Acids Res 33: 6435
- PubMed: 16282592
- DOI: https://doi.org/10.1093/nar/gki945
- Primary Citation of Related Structures:
2C35 - PubMed Abstract:
The Rpb4 and Rpb7 subunits of eukaryotic RNA polymerase II (RNAP(II)) form a heterodimer that protrudes from the 10-subunit core of the enzyme. We have obtained crystals of the human Rpb4/Rpb7 heterodimer and determined the structure to 2.7 A resolution. The presence of putative RNA-binding domains on the Rpb7 subunit and the position of the heterodimer close to the RNA exit groove in the 12 subunit yeast polymerase complex strongly suggests a role for the heterodimer in binding and stabilizing the nascent RNA transcript. We have complemented the structural analysis with biochemical studies directed at dissecting the RNA-binding properties of the human Rpb4/Rpb7 complex and that of the homologous E/F complex from Methanocaldococcus jannaschii. A number of conserved, solvent-exposed residues in both the human Rpb7 subunit and the archaeal E subunit have been modified by site-directed mutagenesis and the mutants tested for RNA binding by performing electrophoretic mobility shift assays. These studies have identified an elongated surface region on the corresponding face of both subunit E and Rpb7 that is involved in RNA binding. The area spans the nucleic acid binding face of the OB fold, including the B4-B5 loop, but also extends towards the N-terminal domain.
Organizational Affiliation:
Department of Biological Sciences, Imperial College London, SW7 2BW, UK.