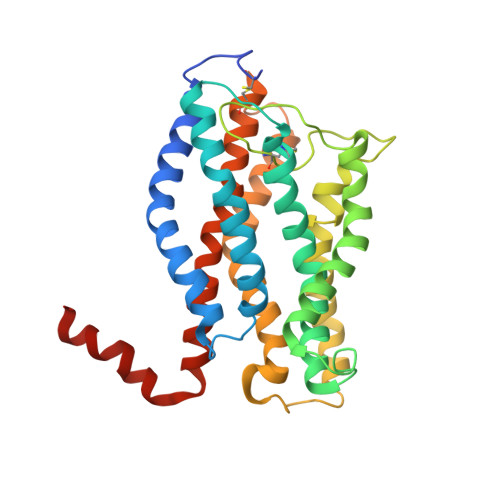
Structure of the chemokine receptor CXCR1 in phospholipid bilayers.
Park, S.H., Das, B.B., Casagrande, F., Tian, Y., Nothnagel, H.J., Chu, M., Kiefer, H., Maier, K., De Angelis, A.A., Marassi, F.M., Opella, S.J.(2012) Nature 491: 779-783
- PubMed: 23086146
- DOI: https://doi.org/10.1038/nature11580
- Primary Citation of Related Structures:
2LNL - PubMed Abstract:
CXCR1 is one of two high-affinity receptors for the CXC chemokine interleukin-8 (IL-8), a major mediator of immune and inflammatory responses implicated in many disorders, including tumour growth. IL-8, released in response to inflammatory stimuli, binds to the extracellular side of CXCR1. The ligand-activated intracellular signalling pathways result in neutrophil migration to the site of inflammation. CXCR1 is a class A, rhodopsin-like G-protein-coupled receptor (GPCR), the largest class of integral membrane proteins responsible for cellular signal transduction and targeted as drug receptors. Despite its importance, the molecular mechanism of CXCR1 signal transduction is poorly understood owing to the limited structural information available. Recent structural determination of GPCRs has advanced by modifying the receptors with stabilizing mutations, insertion of the protein T4 lysozyme and truncations of their amino acid sequences, as well as addition of stabilizing antibodies and small molecules that facilitate crystallization in cubic phase monoolein mixtures. The intracellular loops of GPCRs are crucial for G-protein interactions, and activation of CXCR1 involves both amino-terminal residues and extracellular loops. Our previous nuclear magnetic resonance studies indicate that IL-8 binding to the N-terminal residues is mediated by the membrane, underscoring the importance of the phospholipid bilayer for physiological activity. Here we report the three-dimensional structure of human CXCR1 determined by NMR spectroscopy. The receptor is in liquid crystalline phospholipid bilayers, without modification of its amino acid sequence and under physiological conditions. Features important for intracellular G-protein activation and signal transduction are revealed. The structure of human CXCR1 in a lipid bilayer should help to facilitate the discovery of new compounds that interact with GPCRs and combat diseases such as breast cancer.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, California 92093-0307, USA.