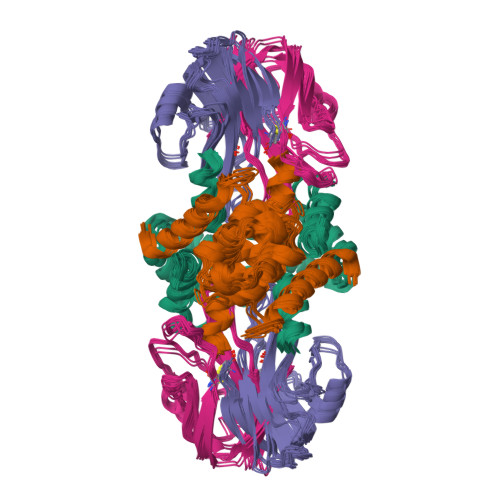
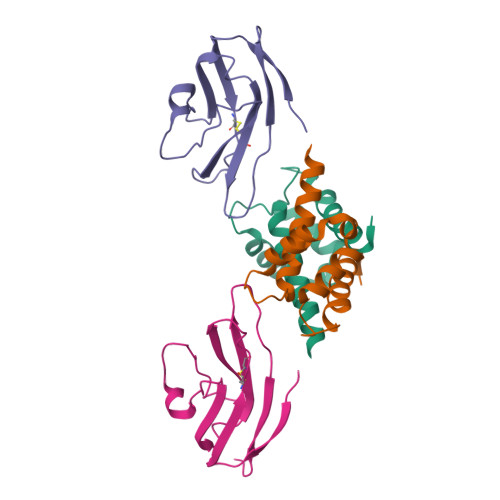
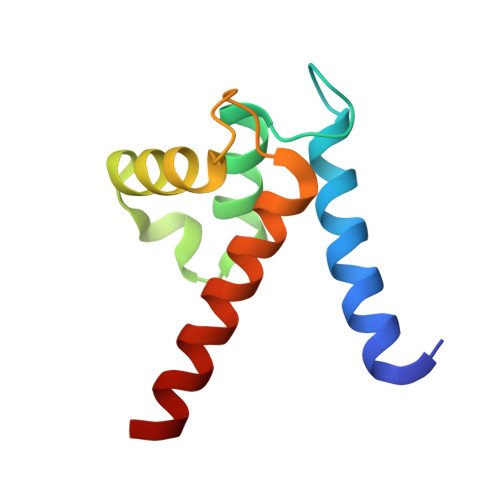
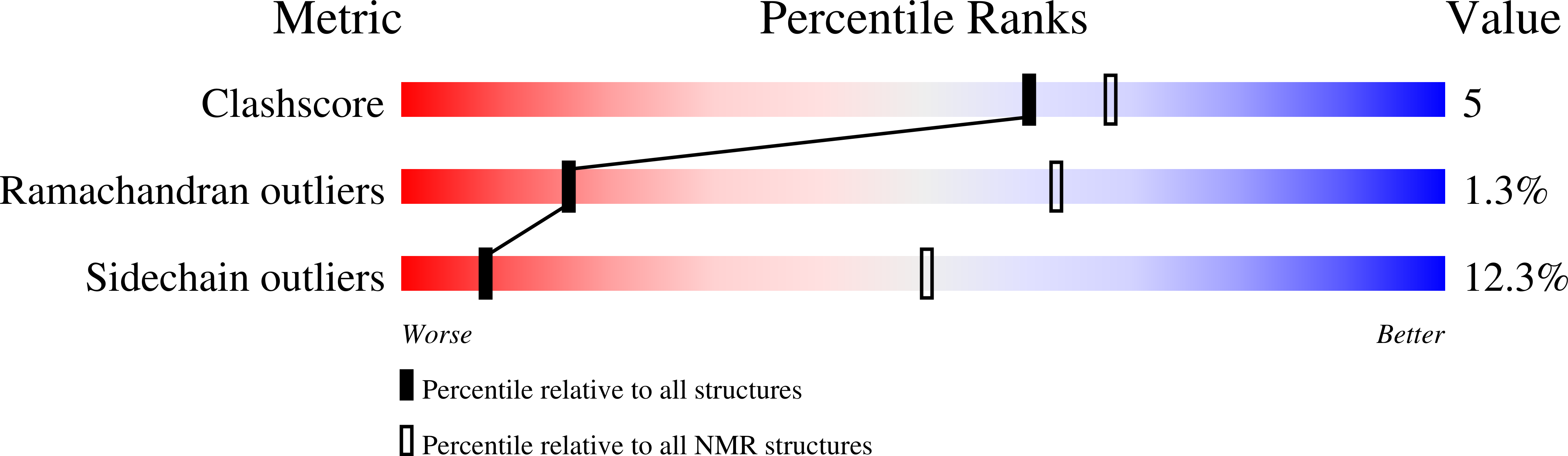Interaction of the S100A6 mutant (C3S) with the V domain of the receptor for advanced glycation end products (RAGE).
Mohan, S.K., Gupta, A.A., Yu, C.(2013) Biochem Biophys Res Commun 434: 328-333
- PubMed: 23537648
- DOI: https://doi.org/10.1016/j.bbrc.2013.03.049
- Primary Citation of Related Structures:
2M1K - PubMed Abstract:
S100A6 is involved in several vital biological functions, such as calcium sensing and cell proliferation. It is a homodimeric protein that belongs to the S100 protein family. The receptor for advanced glycation end products (RAGE) has been shown to play a role in the progression of various disease conditions, such as diabetes and immune/inflammatory disorders. Information regarding the association of RAGE with S100 proteins at a molecular level is useful to understand the diversity of the RAGE signaling pathways. In this report, biomolecular NMR techniques were utilized for the resonance assignment of the C3S mutation in human S100A6 and characterizing its interaction with the RAGE V domain. Further binding affinity between S100A6m and the RAGE V domain was determined by isothermal titration calorimetric studies. HADDOCK was used to generate a heterotetramer model of the S100A6m-RAGE V domain complex. This model provides an important insights into the S100-RAGE cellular signaling pathway.
Organizational Affiliation:
Department of Chemistry, National Tsing Hua University, Hsinchu, Taiwan. mohansepuri@gmail.com