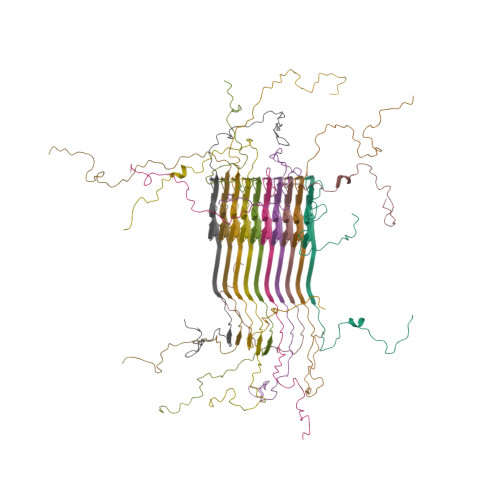
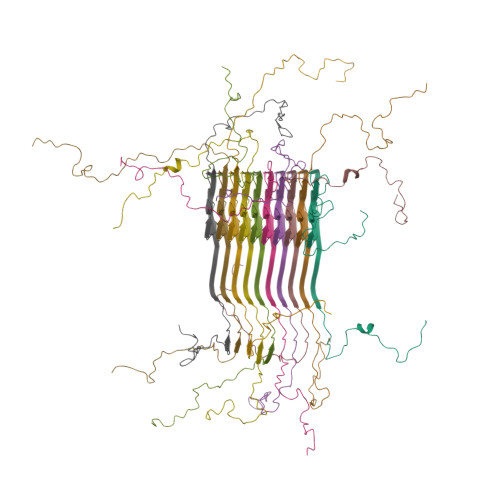
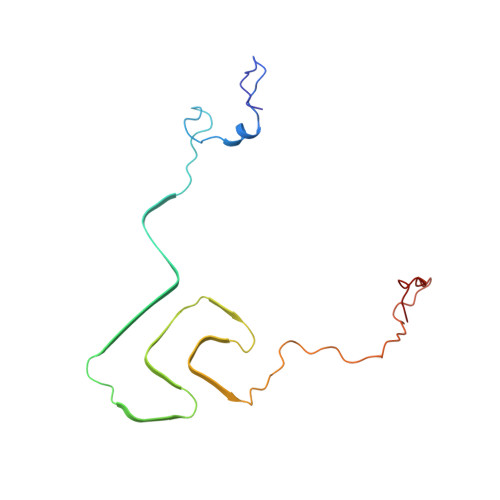
Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein.
Tuttle, M.D., Comellas, G., Nieuwkoop, A.J., Covell, D.J., Berthold, D.A., Kloepper, K.D., Courtney, J.M., Kim, J.K., Barclay, A.M., Kendall, A., Wan, W., Stubbs, G., Schwieters, C.D., Lee, V.M., George, J.M., Rienstra, C.M.(2016) Nat Struct Mol Biol 23: 409-415
- PubMed: 27018801
- DOI: https://doi.org/10.1038/nsmb.3194
- Primary Citation of Related Structures:
2N0A - PubMed Abstract:
Misfolded α-synuclein amyloid fibrils are the principal components of Lewy bodies and neurites, hallmarks of Parkinson's disease (PD). We present a high-resolution structure of an α-synuclein fibril, in a form that induces robust pathology in primary neuronal culture, determined by solid-state NMR spectroscopy and validated by EM and X-ray fiber diffraction. Over 200 unique long-range distance restraints define a consensus structure with common amyloid features including parallel, in-register β-sheets and hydrophobic-core residues, and with substantial complexity arising from diverse structural features including an intermolecular salt bridge, a glutamine ladder, close backbone interactions involving small residues, and several steric zippers stabilizing a new orthogonal Greek-key topology. These characteristics contribute to the robust propagation of this fibril form, as supported by the structural similarity of early-onset-PD mutants. The structure provides a framework for understanding the interactions of α-synuclein with other proteins and small molecules, to aid in PD diagnosis and treatment.
Organizational Affiliation:
Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois, USA.