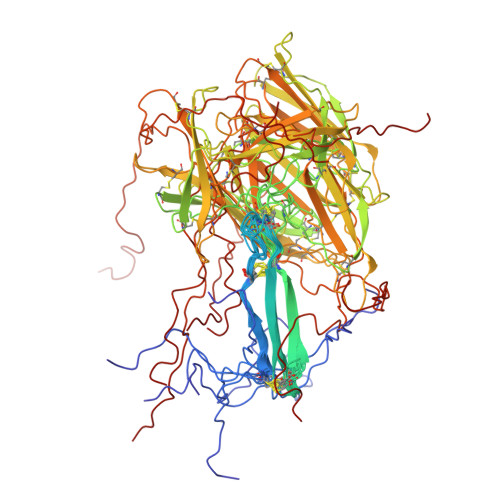
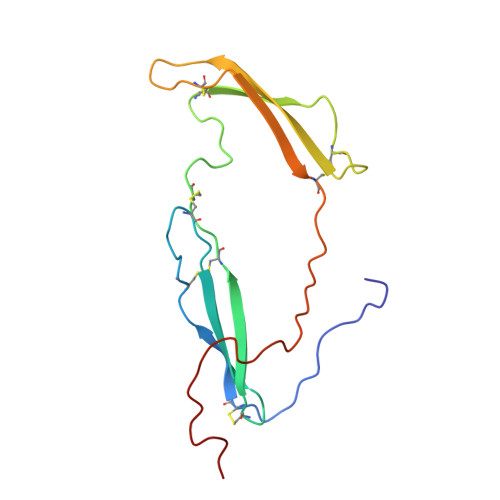
Structural studies reveal an important role for the pleiotrophin C-terminus in mediating interactions with chondroitin sulfate.
Ryan, E., Shen, D., Wang, X.(2016) FEBS J 283: 1488-1503
- PubMed: 26896299
- DOI: https://doi.org/10.1111/febs.13686
- Primary Citation of Related Structures:
2N6F - PubMed Abstract:
Pleiotrophin (PTN) is a potent glycosaminoglycan-binding cytokine that is important in neural development, angiogenesis and tissue regeneration. Much of its activity is attributed to its interactions with the chondroitin sulfate (CS) proteoglycan, receptor type protein tyrosine phosphatase ζ (PTPRZ). However, there is little high resolution structural information on the interactions between PTN and CS, nor is it clear why the C-terminal tail of PTN is necessary for signaling through PTPRZ, even though it does not contribute to heparin binding. We determined the first structure of PTN and analyzed its interactions with CS. Our structure shows that PTN possesses large basic surfaces on both of its structured domains and also that residues in the hinge segment connecting the domains have significant contacts with the C-terminal domain. Our analysis of PTN-CS interactions showed that the C-terminal tail of PTN is essential for maintaining stable interactions with chondroitin sulfate A, the type of CS commonly found on PTPRZ. These results offer the first possible explanation of why truncated PTN missing the C-terminal tail is unable to signal through PTPRZ. NMR analysis of the interactions of PTN with CS revealed that the C-terminal domain and hinge of PTN make up the major CS-binding site in PTN, and that removal of the C-terminal tail weakened the affinity of the site for CSA but not for other high sulfation density CS. Coordinates of the ensemble of ten PTN structures have been deposited in RCSB under accession number 2n6f. Chemical shifts assignments and structural constraints have been deposited in BMRB under accession number 25762.
Organizational Affiliation:
School of Molecular Sciences, Arizona State University, Tempe, AZ, USA.