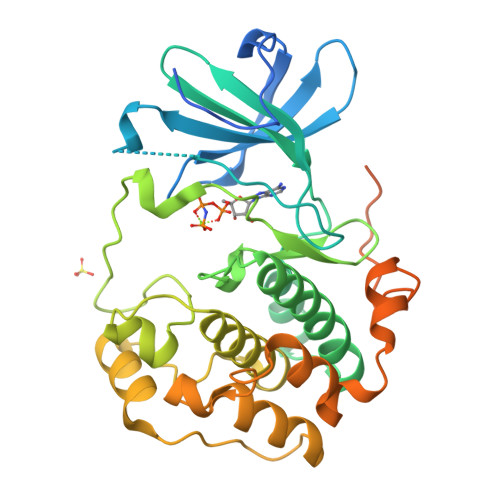
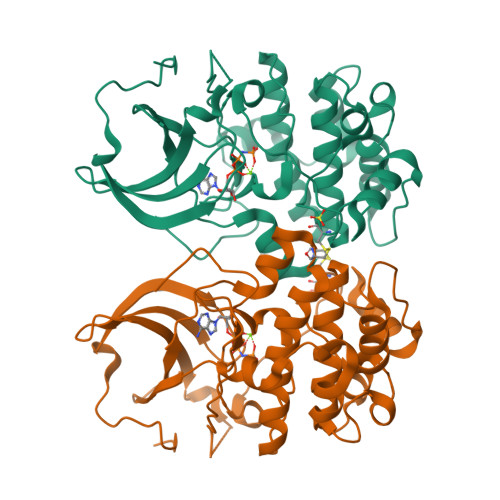
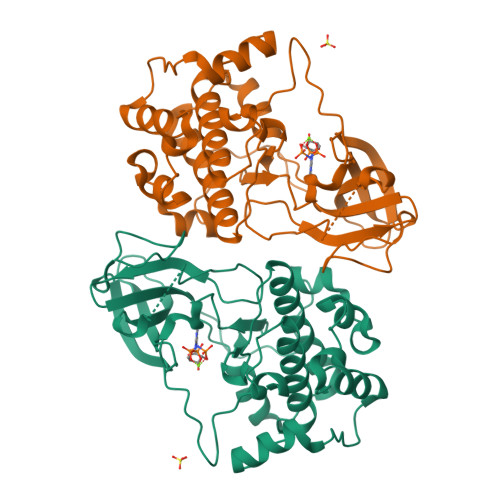
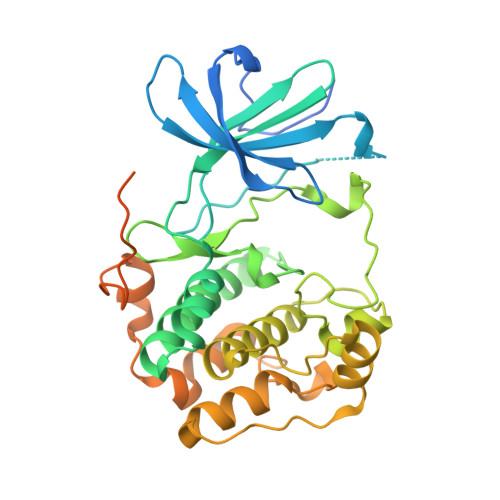
Crystal structure of the kinase domain of serum and glucocorticoid-regulated kinase 1 in complex with AMP PNP.
Zhao, B., Lehr, R., Smallwood, A.M., Ho, T.F., Maley, K., Randall, T., Head, M.S., Koretke, K.K., Schnackenberg, C.G.(2007) Protein Sci 16: 2761-2769
- PubMed: 17965184
- DOI: https://doi.org/10.1110/ps.073161707
- Primary Citation of Related Structures:
2R5T - PubMed Abstract:
Serum and glucocorticoid-regulated kinase 1 (SGK1) is a serine/threonine protein kinase of the AGC family which participates in the control of epithelial ion transport and is implicated in proliferation and apoptosis. We report here the 1.9 A crystal structure of the catalytic domain of inactive human SGK1 in complex with AMP-PNP. SGK1 exists as a dimer formed by two intermolecular disulfide bonds between Cys258 in the activation loop and Cys193. Although most of the SGK1 structure closely resembles the common protein kinase fold, the structure around the active site is unique when compared to most protein kinases. The alphaC helix is not present in this inactive form of SGK1 crystal structure; instead, the segment corresponding to the C helix forms a beta-strand that is stabilized by the N-terminal segment of the activation loop through a short antiparallel beta-sheet. Since the differences from other kinases occur around the ATP binding site, this structure can provide valuable insight into the design of selective and highly potent ATP-competitive inhibitors of SGK1 kinase.
Organizational Affiliation:
Department of Computational and Structural Chemistry, GlaxoSmithKline, King of Prussia, Pennsylvania 19406, USA. baoguang.zhao@gsk.com