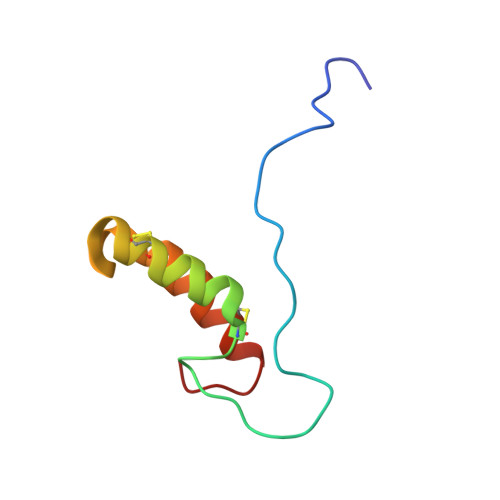
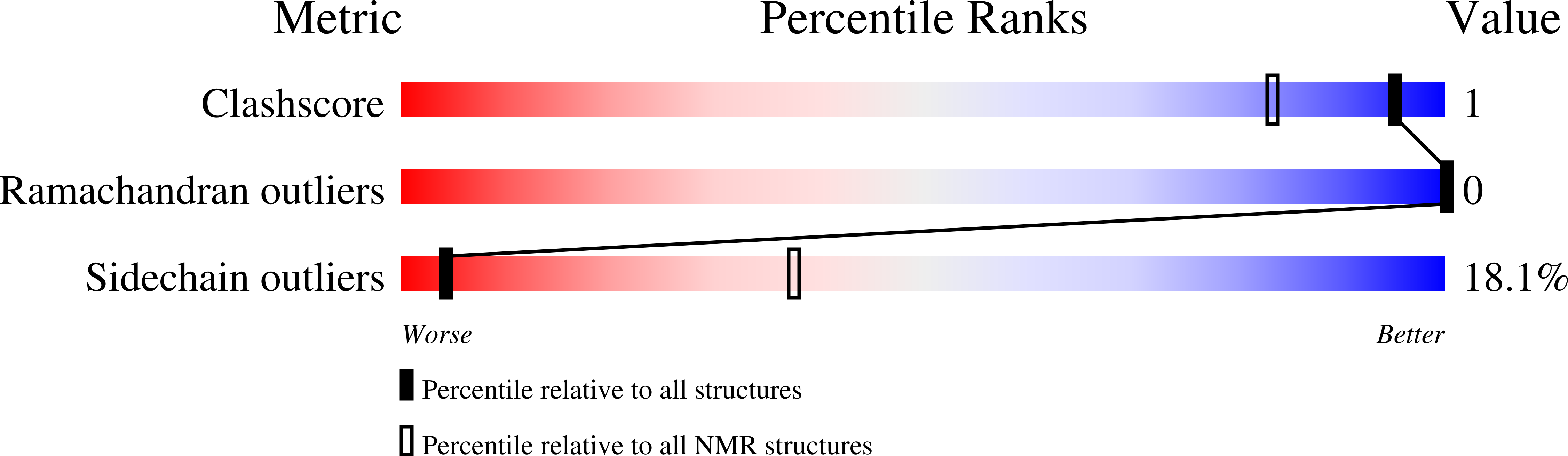A structural-dynamical characterization of human cox17
Banci, L., Bertini, I., Ciofi-Baffoni, S., Janicka, A., Martinelli, M., Kozlowski, H., Palumaa, P.(2008) J Biol Chem 283: 7912-7920
- PubMed: 18093982
- DOI: https://doi.org/10.1074/jbc.M708016200
- Primary Citation of Related Structures:
2RN9, 2RNB - PubMed Abstract:
Human Cox17 is a key mitochondrial copper chaperone responsible for supplying copper ions, through the assistance of Sco1, Sco2, and Cox11, to cytochrome c oxidase, the terminal enzyme of the mitochondrial energy transducing respiratory chain. A structural and dynamical characterization of human Cox17 in its various functional metallated and redox states is presented here. The NMR solution structure of the partially oxidized Cox17 (Cox17(2S-S)) consists of a coiled coil-helix-coiled coil-helix domain stabilized by two disulfide bonds involving Cys(25)-Cys(54) and Cys(35)-Cys(44), preceded by a flexible and completely unstructured N-terminal tail. In human Cu(I)Cox17(2S-S) the copper(I) ion is coordinated by the sulfurs of Cys(22) and Cys(23), and this is the first example of a Cys-Cys binding motif in copper proteins. Copper(I) binding as well as the formation of a third disulfide involving Cys(22) and Cys(23) cause structural and dynamical changes only restricted to the metal-binding region. Redox properties of the disulfides of human Cox17, here investigated, strongly support the current hypothesis that the unstructured fully reduced Cox17 protein is present in the cytoplasm and enters the intermembrane space (IMS) where is then oxidized by Mia40 to Cox17(2S-S), thus becoming partially structured and trapped into the IMS. Cox17(2S-S) is the functional species in the IMS, it can bind only one copper(I) ion and is then ready to enter the pathway of copper delivery to cytochrome c oxidase. The copper(I) form of Cox17(2S-S) has features specific for copper chaperones.
Organizational Affiliation:
Magnetic Resonance Center Centro Risonanze Magnetiche (CERM) and Department of Chemistry, University of Florence, Via Luigi Sacconi 6, 50019, Sesto Fiorentino, Florence, Italy.