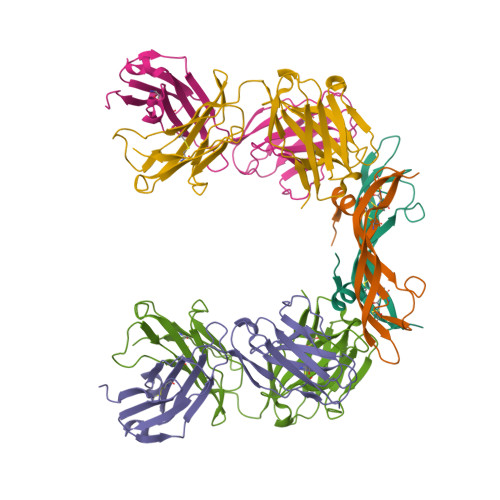
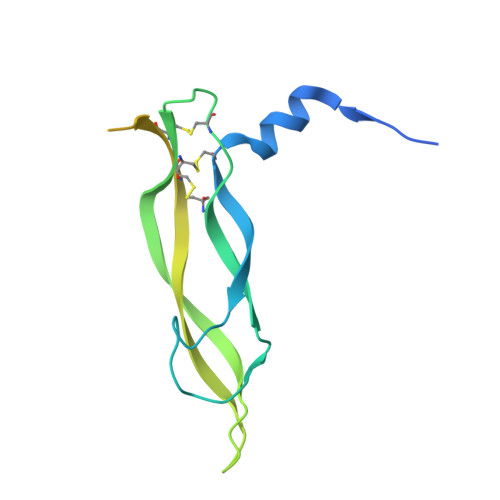
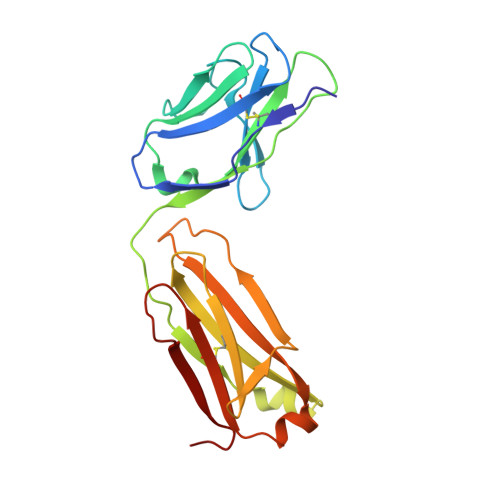
Crystal Structure of Vascular Endothelial Growth Factor-B in Complex with a Neutralising Antibody Fab Fragment.
Leonard, P., Scotney, P.D., Jabeen, T., Iyer, S., Fabri, L.J., Nash, A.D., Acharya, K.R.(2008) J Mol Biology 384: 1203
- PubMed: 18930733
- DOI: https://doi.org/10.1016/j.jmb.2008.09.076
- Primary Citation of Related Structures:
2VWE - PubMed Abstract:
Vascular endothelial growth factor (VEGF) B effects blood vessel formation by binding to VEGF receptor 1. To study the specifics of the biological profile of VEGF-B in both physiological and pathological angiogenesis, a neutralising anti-VEGF-B antibody (2H10) that functions by inhibiting the binding of VEGF-B to VEGF receptor 1 was developed. Here, we present the structural features of the 'highly ordered' interaction of the Fab fragment of this antibody (Fab-2H10) with VEGF-B. Two molecules of Fab-2H10 bind to symmetrical binding sites located at each pole of the VEGF-B homodimer, giving a unique U-shaped topology to the complex that has not been previously observed in the VEGF family. VEGF-B residues essential for binding to the antibody are contributed by both monomers of the cytokine. Our detailed analysis reveals that the neutralising effect of the antibody occurs by virtue of the steric hindrance of the receptor-binding interface. These findings suggest that functional complementarity between VEGF-B and 2H10 can be harnessed both in analysing the therapeutic potential of VEGF-B and as an antagonist of receptor activation.
Organizational Affiliation:
Department of Biology and Biochemistry, University of Bath, Claverton Down, Bath BA2 7AY, UK.