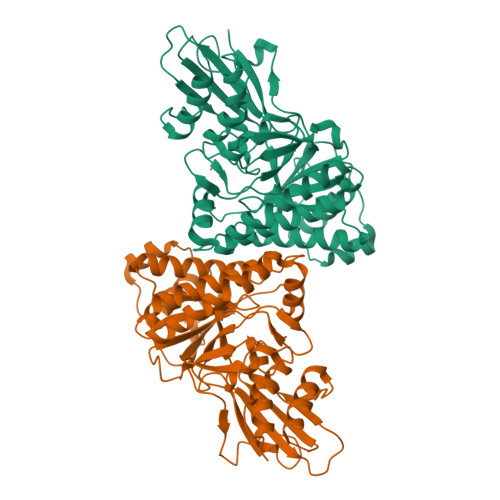
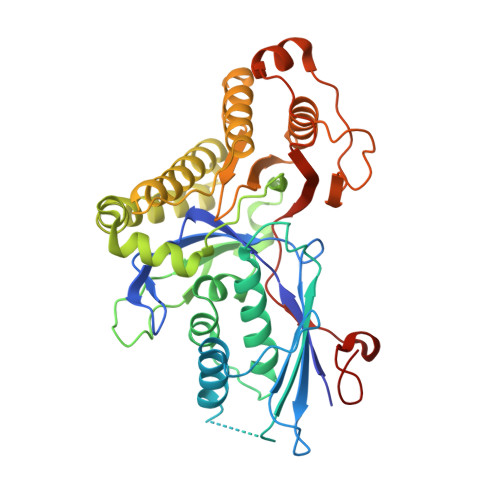
Human mevalonate diphosphate decarboxylase: characterization, investigation of the mevalonate diphosphate binding site, and crystal structure.
Voynova, N.E., Fu, Z., Battaile, K.P., Herdendorf, T.J., Kim, J.J., Miziorko, H.M.(2008) Arch Biochem Biophys 480: 58-67
- PubMed: 18823933
- DOI: https://doi.org/10.1016/j.abb.2008.08.024
- Primary Citation of Related Structures:
3D4J - PubMed Abstract:
Expression in Escherichia coli of his-tagged human mevalonate diphosphate decarboxylase (hMDD) has expedited enzyme isolation, characterization, functional investigation of the mevalonate diphosphate binding site, and crystal structure determination (2.4A resolution). hMDD exhibits V(max)=6.1+/-0.5 U/mg; K(m) for ATP is 0.69+/-0.07 mM and K(m) for (R,S) mevalonate diphosphate is 28.9+/-3.3 microM. Conserved polar residues predicted to be in the hMDD active site were mutated to test functional importance. R161Q exhibits a approximately 1000-fold diminution in specific activity, while binding the fluorescent substrate analog, TNP-ATP, comparably to wild-type enzyme. Diphosphoglycolyl proline (K(i)=2.3+/-0.3 uM) and 6-fluoromevalonate 5-diphosphate (K(i)=62+/-5 nM) are competitive inhibitors with respect to mevalonate diphosphate. N17A exhibits a V(max)=0.25+/-0.0 2U/mg and a 15-fold inflation in K(m) for mevalonate diphosphate. N17A's K(i) values for diphosphoglycolyl proline and fluoromevalonate diphosphate are inflated (>70-fold and 40-fold, respectively) in comparison with wild-type enzyme. hMDD structure indicates the proximity (2.8A) between R161 and N17, which are located in an interior pocket of the active site cleft. The data suggest the functional importance of R161 and N17 in the binding and orientation of mevalonate diphosphate.
Organizational Affiliation:
Division of Molecular Biology and Biochemistry, University of Missouri-Kansas City, 5007 Rockhill Road, Kansas City, MO 64110, USA.