Structural insight into the activation mechanism of human pancreatic prophospholipase A2
Xu, W., Yi, L., Feng, Y., Chen, L., Liu, J.(2009) J Biological Chem 284: 16659-16666
- PubMed: 19297324
- DOI: https://doi.org/10.1074/jbc.M808029200
- Primary Citation of Related Structures:
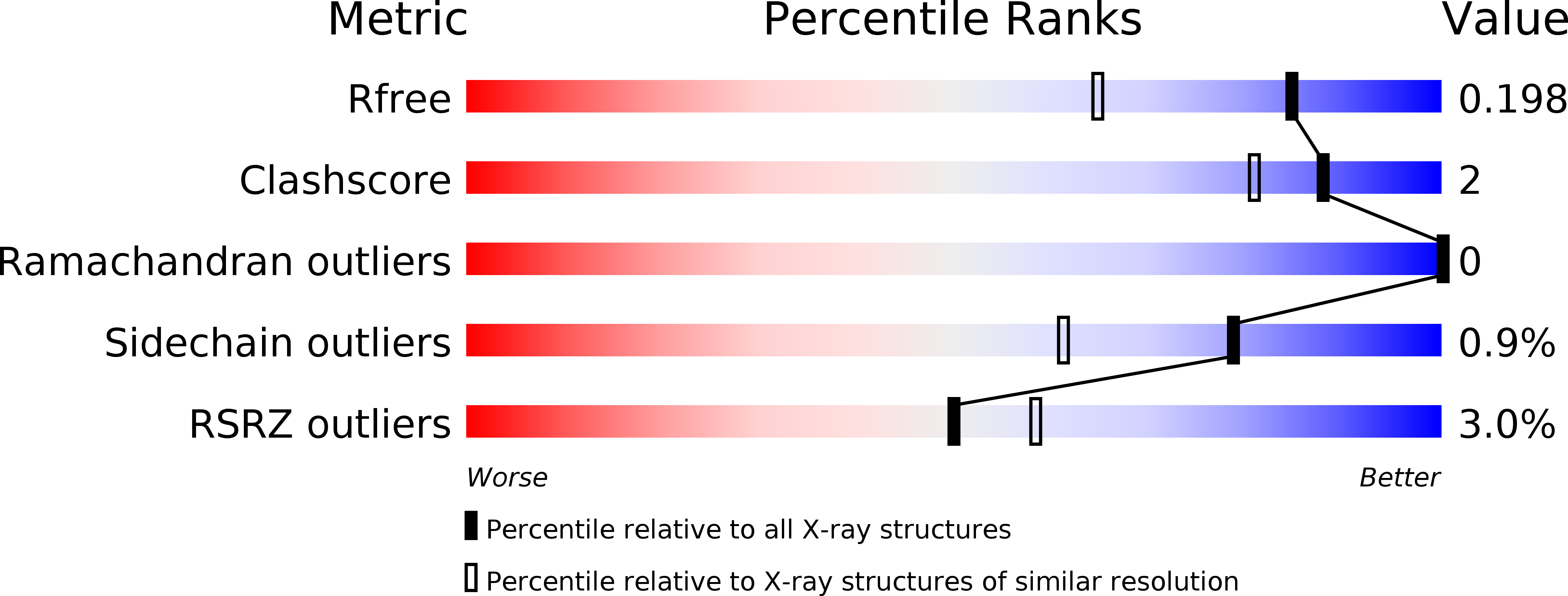
3ELO - PubMed Abstract:
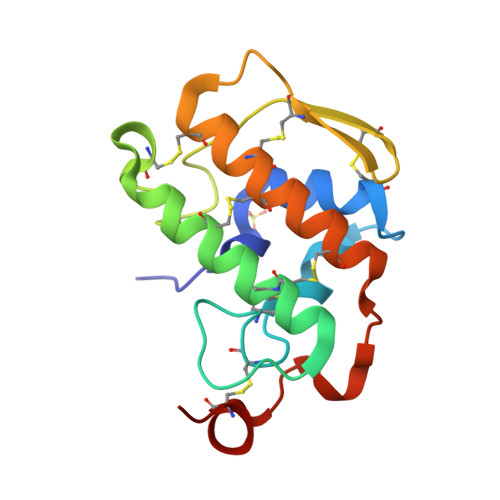
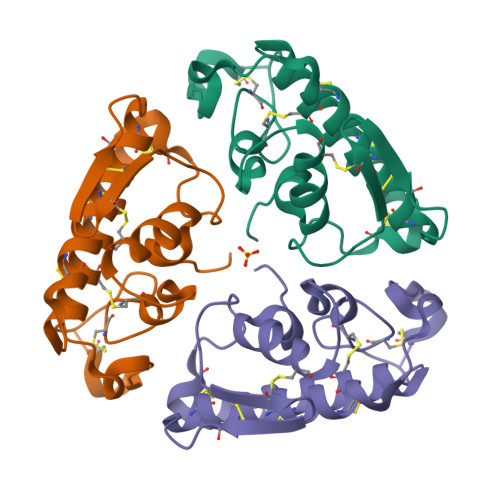
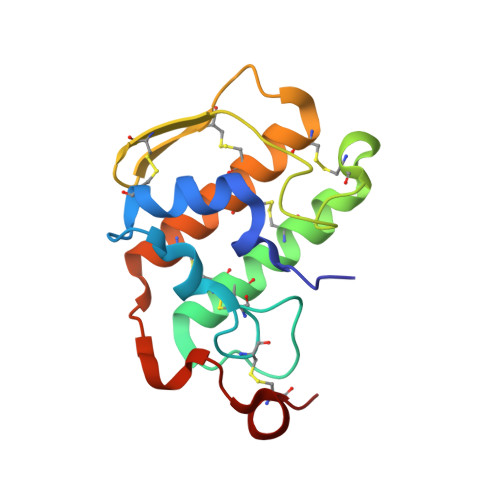
Pancreatic phospholipase A2 (phospholipase A2 group 1B, G1B) belongs to the superfamily of secreted phospholipase A2 (PLA2) enzymes. G1B has been proposed to be a potential target for diseases such as hypertension, obesity, and diabetes. Human pancreatic prophospholipase A2 (pro-hG1B) is activated by cleavage of the first seven-residue propeptide (phospholipase A2 propeptide, PROP). However, questions still remain on the mode of action for pro-hG1B. In this work, we expressed pro-hG1B in Pichia pastoris and determined the crystal structure at 1.55-A resolution. The x-ray structure demonstrates that pro-hG1B forms a trimer. In addition, PROP occupies the catalytic cavity and can be self-cleaved at 37 degrees C. A new membrane-bound surface and activation mechanism are proposed based on the trimeric model of pro-hG1B. We also propose a new autoproteolytic mechanism for pro-hG1B by the reaction triad Asp49-Arg0-Ser(-2) that is similar to the serine protease catalytic triad.
Organizational Affiliation:
From the Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510663, China; Graduate University of Chinese Academy of Sciences, Beijing 100049, China.