Paired octamer rings of retinoschisin suggest a junctional model for cell-cell adhesion in the retina.
Tolun, G., Vijayasarathy, C., Huang, R., Zeng, Y., Li, Y., Steven, A.C., Sieving, P.A., Heymann, J.B.(2016) Proc Natl Acad Sci U S A 113: 5287-5292
- PubMed: 27114531
- DOI: https://doi.org/10.1073/pnas.1519048113
- Primary Citation of Related Structures:
3JD6 - PubMed Abstract:
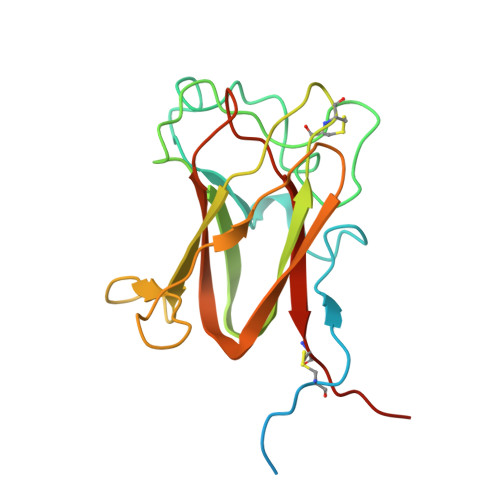
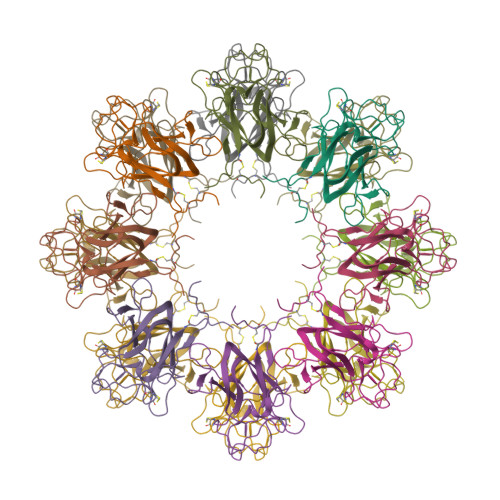
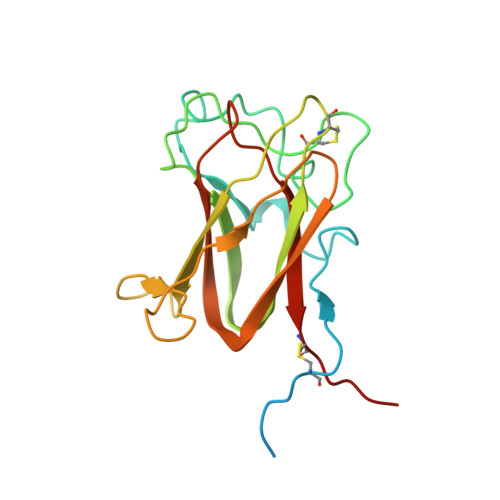
Retinoschisin (RS1) is involved in cell-cell junctions in the retina, but is unique among known cell-adhesion proteins in that it is a soluble secreted protein. Loss-of-function mutations in RS1 lead to early vision impairment in young males, called X-linked retinoschisis. The disease is characterized by separation of inner retinal layers and disruption of synaptic signaling. Using cryo-electron microscopy, we report the structure at 4.1 Å, revealing double octamer rings not observed before. Each subunit is composed of a discoidin domain and a small N-terminal (RS1) domain. The RS1 domains occupy the centers of the rings, but are not required for ring formation and are less clearly defined, suggesting mobility. We determined the structure of the discoidin rings, consistent with known intramolecular and intermolecular disulfides. The interfaces internal to and between rings feature residues implicated in X-linked retinoschisis, indicating the importance of correct assembly. Based on this structure, we propose that RS1 couples neighboring membranes together through octamer-octamer contacts, perhaps modulated by interactions with other membrane components.
Organizational Affiliation:
Laboratory for Structural Biology Research, National Institute of Arthritis, Musculoskeletal, and Skin Diseases, National Institutes of Health, Bethesda, MD 20892; Molecular Control and Genetics Section, Gene Regulation and Chromosome Biology Laboratory, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Frederick, MD 21702;