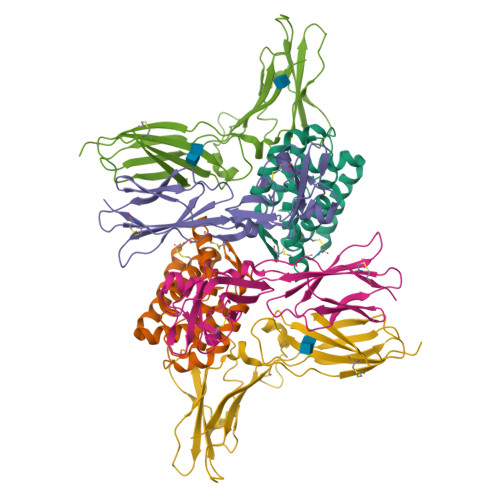
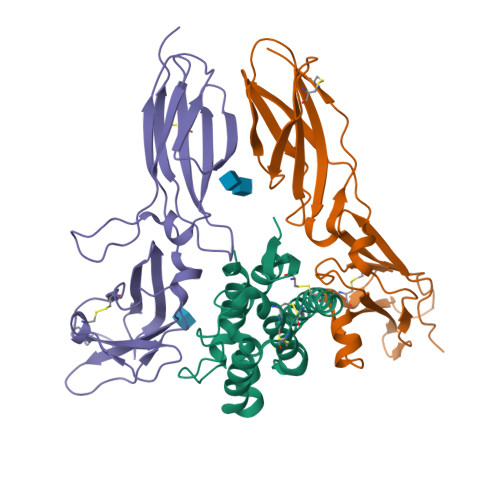
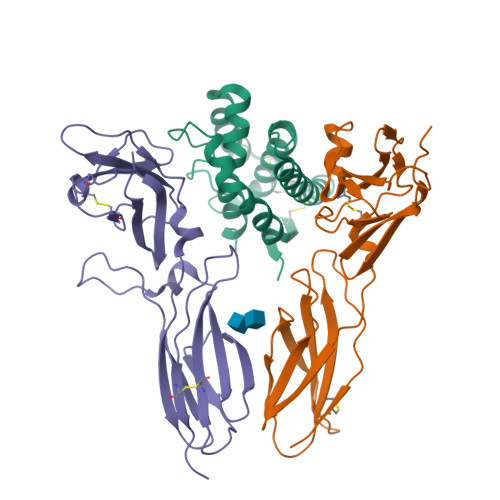
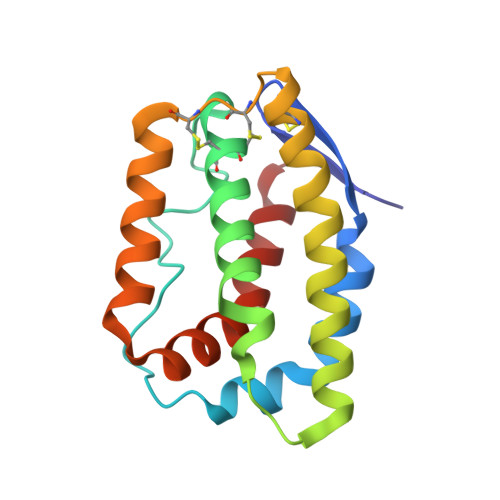
Structural basis for receptor sharing and activation by interleukin-20 receptor-2 (IL-20R2) binding cytokines.
Logsdon, N.J., Deshpande, A., Harris, B.D., Rajashankar, K.R., Walter, M.R.(2012) Proc Natl Acad Sci U S A 109: 12704-12709
- PubMed: 22802649
- DOI: https://doi.org/10.1073/pnas.1117551109
- Primary Citation of Related Structures:
4DOH - PubMed Abstract:
Interleukin 20 (IL-20) is a pleotropic IL-10 family cytokine that protects epithelial surfaces from pathogens. However, dysregulated IL-20 signaling is implicated in several human pathologies including psoriasis, rheumatoid arthritis, atherosclerosis, and osteoporosis. IL-20, and related cytokines IL-19 and IL-24, designated IL-20 subfamily cytokines (IL-20SFCs), induce cellular responses through an IL-20R1/IL-20R2 (type I) receptor heterodimer, whereas IL-20 and IL-24 also signal through the IL-22R1/IL-20R2 (type II) receptor complex. The crystal structure of the IL-20/IL-20R1/IL-20R2 complex reveals how type I and II complexes discriminate cognate from noncognate ligands. The structure also defines how the receptor-cytokine interfaces are affinity tuned to allow distinct signaling through a receptor complex shared by three different ligands. Our results provide unique insights into the complexity of IL-20SFC signaling that may be critical in the design of mechanistic-based inhibitors of IL-20SFC-mediated inflammatory disease.
Organizational Affiliation:
Center for Biophysical Sciences, University of Alabama, Birmingham, AL 35294, USA.