Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Zhang, Q., Qi, S., Xu, M., Yu, L., Tao, Y., Deng, Z., Wu, W., Li, J., Chen, Z., Wong, J.(2013) Cell Res 23: 225-241
- PubMed: 23266887
- DOI: https://doi.org/10.1038/cr.2012.177
- Primary Citation of Related Structures:
4FWE, 4FWF, 4FWJ - PubMed Abstract:
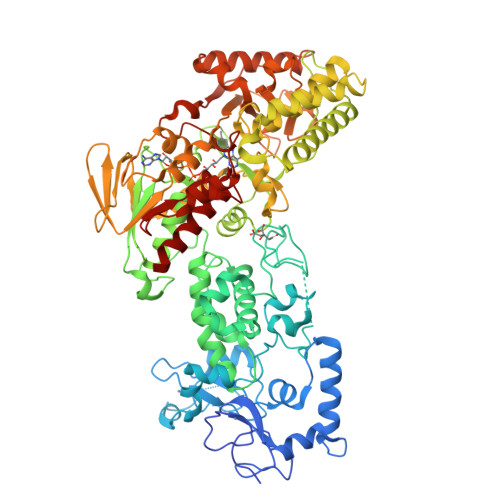
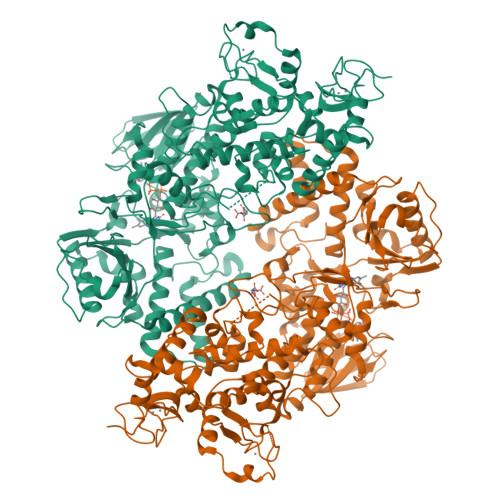
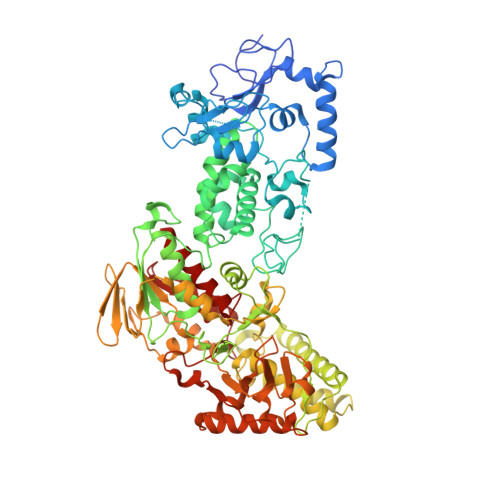
LSD2/AOF1/KDM1b catalyzes demethylation of mono- and di-methylated H3K4 and plays an important role in transcriptional regulation and genomic imprinting. Here, we report the high-resolution crystal structures of apo-LSD2 and LSD2 in complex with a peptide that mimics H3K4me2. Three structural domains of LSD2, namely, the novel N-terminal zinc finger, the centrally located SWIRM domain, and the C-terminal oxidase domain, closely pack together to form a boot-shaped structure. The active site cavity in the oxidase domain is large enough to accommodate several residues of the histone H3 tail and cannot discriminate between the different states of H3K4 methylation. The N-terminal zinc-finger domain, composed of a novel C4H2C2-type zinc finger and a specific CW-type zinc finger, is required for demethylase activity and, surprisingly, the binding of cofactor flavin adenine dinucleotide (FAD). In fact, a relay of extensive interactions through the zinc finger-SWIRM-oxidase domains is required for LSD2 demethylase activity and the binding of FAD. These results reveal a novel mechanism for the zinc finger and SWIRM domains in controlling LSD2 demethylase activity and provide a framework for elucidating the regulation and function of LSD2.
Organizational Affiliation:
State Key Laboratory of Agrobiotechnology, China Agricultural University, Beijing 100193, China.