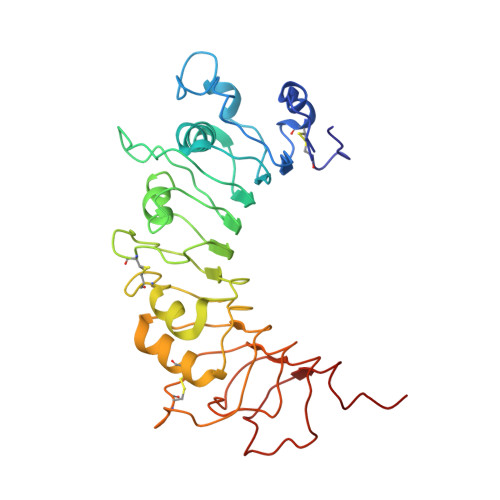
The Crystal Structure of Human Soluble CD14 Reveals a Bent Solenoid with a Hydrophobic Amino-Terminal Pocket.
Kelley, S.L., Lukk, T., Nair, S.K., Tapping, R.I.(2013) J Immunol 190: 1304-1311
- PubMed: 23264655
- DOI: https://doi.org/10.4049/jimmunol.1202446
- Primary Citation of Related Structures:
4GLP - PubMed Abstract:
Human monocyte differentiation Ag CD14 is a pattern recognition receptor that enhances innate immune responses to infection by sensitizing host cells to bacterial LPS (endotoxin), lipoproteins, lipoteichoic acid, and other acylated microbial products. CD14 physically delivers these lipidated microbial products to various TLR signaling complexes that subsequently induce intracellular proinflammatory signaling cascades upon ligand binding. The ensuing cellular responses are usually protective to the host but can also result in host fatality through sepsis. In this work, we have determined the x-ray crystal structure of human CD14. The structure reveals a bent solenoid typical of leucine-rich repeat proteins with an amino-terminal pocket that presumably binds acylated ligands including LPS. Comparison of human and mouse CD14 structures shows great similarity in overall protein fold. However, compared with mouse CD14, human CD14 contains an expanded pocket and alternative rim residues that are likely to be important for LPS binding and cell activation. The x-ray crystal structure of human CD14 presented in this article may foster additional ligand-bound structural studies, virtual docking studies, and drug design efforts to mitigate LPS-induced sepsis and other inflammatory diseases.
Organizational Affiliation:
Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.