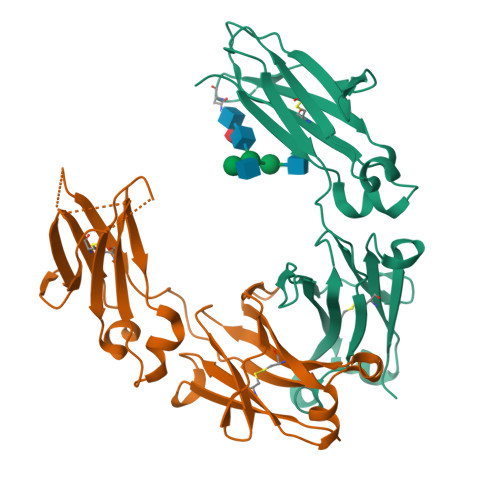
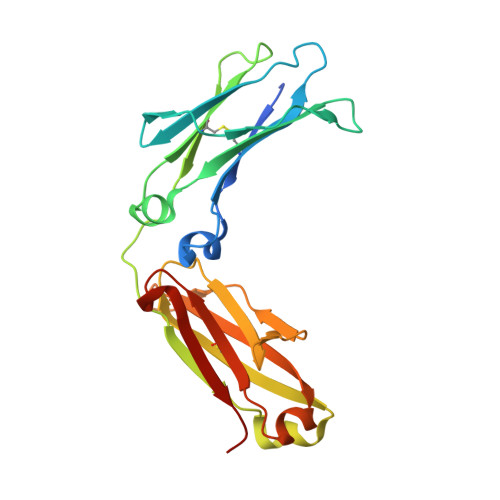
IgG2 Fc structure and the dynamic features of the IgG CH2-CH3 interface.
Teplyakov, A., Zhao, Y., Malia, T.J., Obmolova, G., Gilliland, G.L.(2013) Mol Immunol 56: 131-139
- PubMed: 23628091
- DOI: https://doi.org/10.1016/j.molimm.2013.03.018
- Primary Citation of Related Structures:
4HAF, 4HAG - PubMed Abstract:
The analyses of two human IgG2 Fc structures, determined in different crystal forms, and the comparison with IgG1 Fc structures reveals molecular features that are involved in accommodating and stabilizing structural conformations. In the IgG2 Fc structures relative positions of the CH2 domains with respect to the CH3 domains vary significantly from those observed for IgG1 Fc structures in similar unit cells. The analysis reveals that the movement of the CH2 domain in all of the Fc structures results from a pivoting around a highly conserved ball-and-socket-like joint in which the CH2 L251 side chain (the ball) interacts with a pocket (the socket) formed by CH3 M428, H429, E430, and H435. Despite the change in positioning of the CH2 and CH3 domains, conserved hydrogen bonds and electrostatic interactions are retained, stabilizing the Fc domain interface. In the high resolution IgG2 and IgG1 Fc structures the position and number of water molecules, and water networks bridging the two domains differ significantly because of the difference in positions of CH2 relative to CH3. At the domain interface, only CH2 T339 in IgG2 differs from alanine found in IgG1 and IgG4. This residue's side chain influences the water structure at the interface by interacting either directly or through a bridging water molecule with D376 in the CH3 BC loop. Thus, the analyses of the IgG2 Fc structures and their comparisons with IgG1 Fc structures reveals similar, but distinctly different dynamic CH2-CH3 interfaces that can accommodate a wide range of CH2-CH3 conformations, that in conjunction with the amino acid residues in the hinge region, may influence FcγR effector function profiles.
Organizational Affiliation:
Biologics Research, Biotechnology Center of Excellence, Janssen R&D LLC, Spring House, PA 19477, United States. ateplyak@its.jnj.com