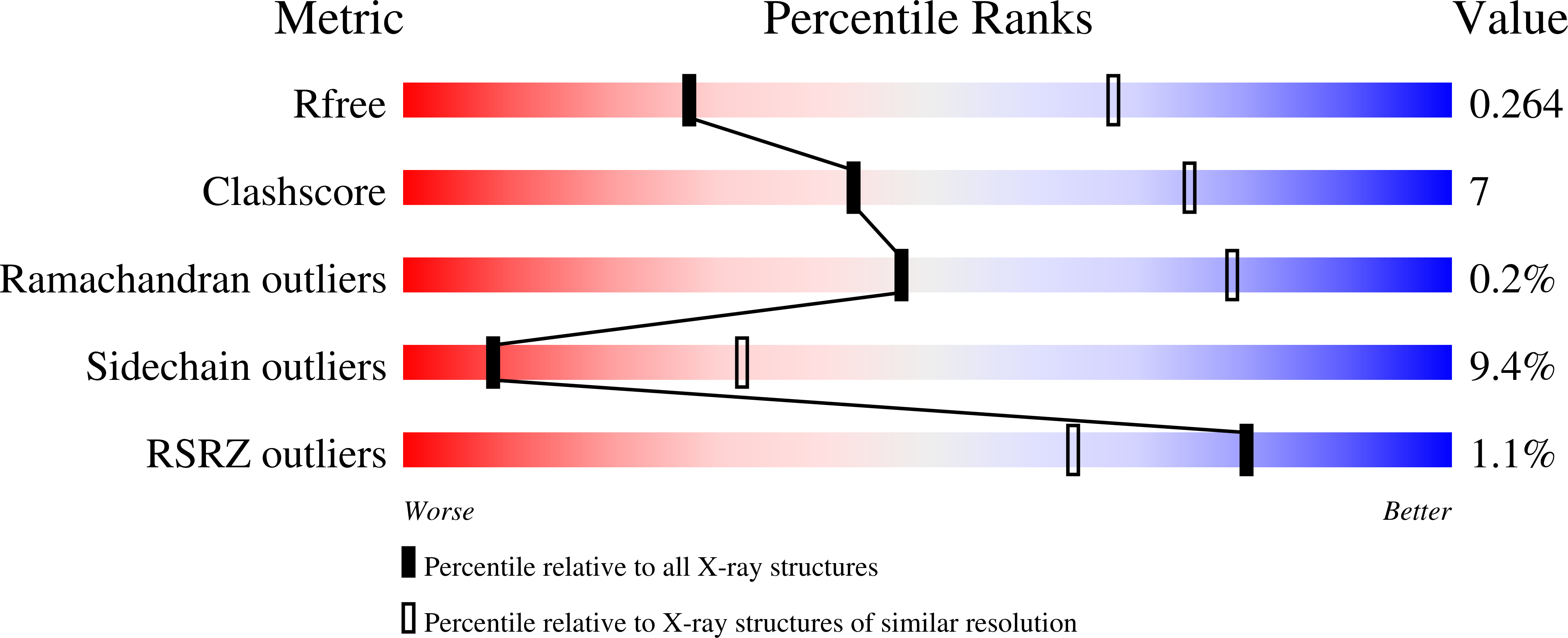
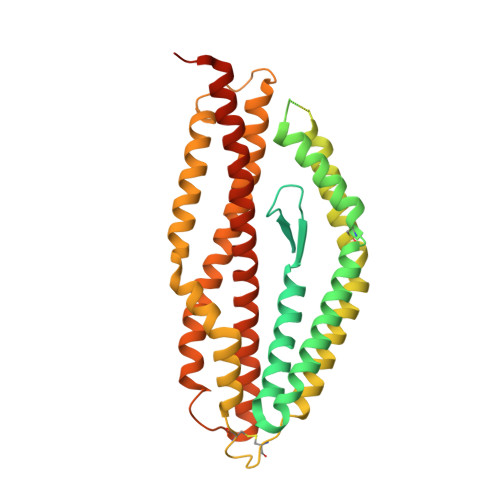
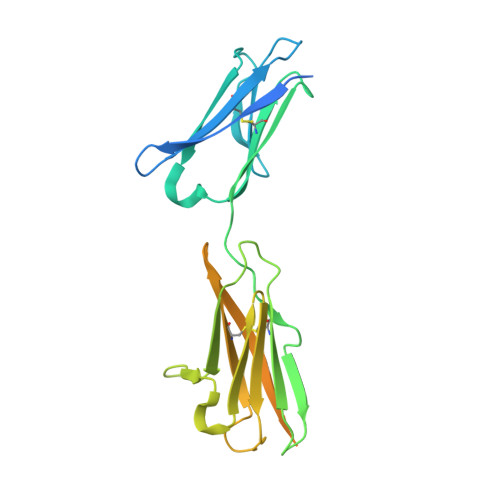
Structure of malaria invasion protein RH5 with erythrocyte basigin and blocking antibodies.
Wright, K.E., Hjerrild, K.A., Bartlett, J., Douglas, A.D., Jin, J., Brown, R.E., Illingworth, J.J., Ashfield, R., Clemmensen, S.B., de Jongh, W.A., Draper, S.J., Higgins, M.K.(2014) Nature 515: 427-430
- PubMed: 25132548
- DOI: https://doi.org/10.1038/nature13715
- Primary Citation of Related Structures:
4U0Q, 4U0R, 4U1G - PubMed Abstract:
Invasion of host erythrocytes is essential to the life cycle of Plasmodium parasites and development of the pathology of malaria. The stages of erythrocyte invasion, including initial contact, apical reorientation, junction formation, and active invagination, are directed by coordinated release of specialized apical organelles and their parasite protein contents. Among these proteins, and central to invasion by all species, are two parasite protein families, the reticulocyte-binding protein homologue (RH) and erythrocyte-binding like proteins, which mediate host-parasite interactions. RH5 from Plasmodium falciparum (PfRH5) is the only member of either family demonstrated to be necessary for erythrocyte invasion in all tested strains, through its interaction with the erythrocyte surface protein basigin (also known as CD147 and EMMPRIN). Antibodies targeting PfRH5 or basigin efficiently block parasite invasion in vitro, making PfRH5 an excellent vaccine candidate. Here we present crystal structures of PfRH5 in complex with basigin and two distinct inhibitory antibodies. PfRH5 adopts a novel fold in which two three-helical bundles come together in a kite-like architecture, presenting binding sites for basigin and inhibitory antibodies at one tip. This provides the first structural insight into erythrocyte binding by the Plasmodium RH protein family and identifies novel inhibitory epitopes to guide design of a new generation of vaccines against the blood-stage parasite.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, UK.