Inter-Domain Communication of Human Cystathionine Beta Synthase: Structural Basis of S-Adenosyl-L-Methionine Activation.
Mccorvie, T.J., Kopec, J., Hyung, S., Fitzpatrick, F., Feng, X., Termine, D., Strain-Damerell, C., Vollmar, M., Fleming, J., Janz, J.M., Bulawa, C., Yue, W.W.(2014) J Biological Chem 289: 36018
- PubMed: 25336647
- DOI: https://doi.org/10.1074/jbc.M114.610782
- Primary Citation of Related Structures:
4COO, 4UUU - PubMed Abstract:
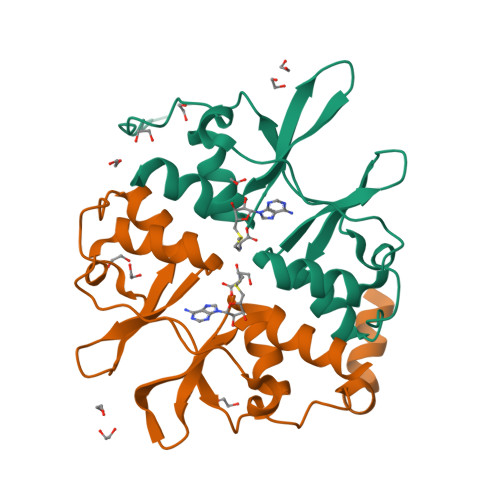
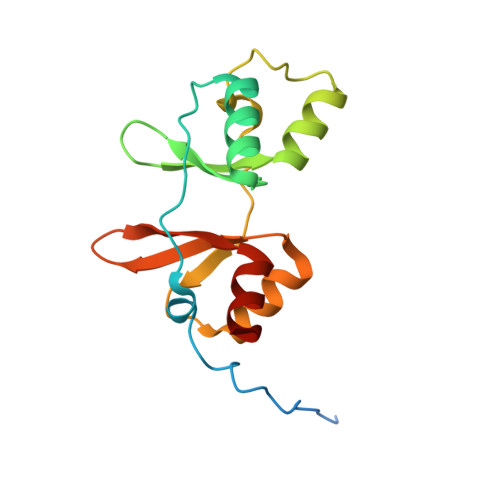
Cystathionine β-synthase (CBS) is a key enzyme in sulfur metabolism, and its inherited deficiency causes homocystinuria. Mammalian CBS is modulated by the binding of S-adenosyl-l-methionine (AdoMet) to its regulatory domain, which activates its catalytic domain. To investigate the underlying mechanism, we performed x-ray crystallography, mutagenesis, and mass spectrometry (MS) on human CBS. The 1.7 Å structure of a AdoMet-bound CBS regulatory domain shows one AdoMet molecule per monomer, at the interface between two constituent modules (CBS-1, CBS-2). AdoMet binding is accompanied by a reorientation between the two modules, relative to the AdoMet-free basal state, to form interactions with AdoMet via residues verified by mutagenesis to be important for AdoMet binding (Phe(443), Asp(444), Gln(445), and Asp(538)) and for AdoMet-driven inter-domain communication (Phe(443), Asp(538)). The observed structural change is further supported by ion mobility MS, showing that as-purified CBS exists in two conformational populations, which converged to one in the presence of AdoMet. We therefore propose that AdoMet-induced conformational change alters the interface and arrangement between the catalytic and regulatory domains within the CBS oligomer, thereby increasing the accessibility of the enzyme active site for catalysis.
Organizational Affiliation:
From the Structural Genomics Consortium, Nuffield Department of Clinical Medicine, University of Oxford, Oxford OX3 7DQ, United Kingdom.