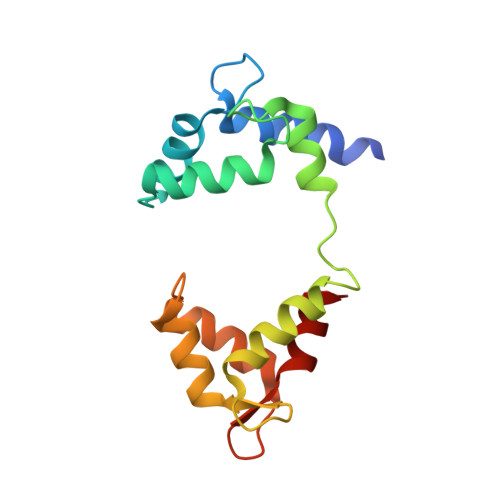
Structural Basis of a Kv7.1 Potassium Channel Gating Module: Studies of the Intracellular C-Terminal Domain in Complex with Calmodulin.
Sachyani, D., Dvir, M., Strulovich, R., Tria, G., Tobelaim, W., Peretz, A., Pongs, O., Svergun, D., Attali, B., Hirsch, J.A.(2014) Structure 22: 1582
- PubMed: 25441029
- DOI: https://doi.org/10.1016/j.str.2014.07.016
- Primary Citation of Related Structures:
4UMO, 4V0C - PubMed Abstract:
Kv7 channels tune neuronal and cardiomyocyte excitability. In addition to the channel membrane domain, they also have a unique intracellular C-terminal (CT) domain, bound constitutively to calmodulin (CaM). This CT domain regulates gating and tetramerization. We investigated the structure of the membrane proximal CT module in complex with CaM by X-ray crystallography. The results show how the CaM intimately hugs a two-helical bundle, explaining many channelopathic mutations. Structure-based mutagenesis of this module in the context of concatemeric tetramer channels and functional analysis along with in vitro data lead us to propose that one CaM binds to one individual protomer, without crosslinking subunits and that this configuration is required for proper channel expression and function. Molecular modeling of the CT/CaM complex in conjunction with small-angle X-ray scattering suggests that the membrane proximal region, having a rigid lever arm, is a critical gating regulator.