Specific GFP-binding artificial proteins ( alpha Rep): a new tool for in vitro to live cell applications.
Chevrel, A., Urvoas, A., de la Sierra-Gallay, I.L., Aumont-Nicaise, M., Moutel, S., Desmadril, M., Perez, F., Gautreau, A., van Tilbeurgh, H., Minard, P., Valerio-Lepiniec, M.(2015) Biosci Rep 35
- PubMed: 26182430
- DOI: https://doi.org/10.1042/BSR20150080
- Primary Citation of Related Structures:
4XL5, 4XVP - PubMed Abstract:
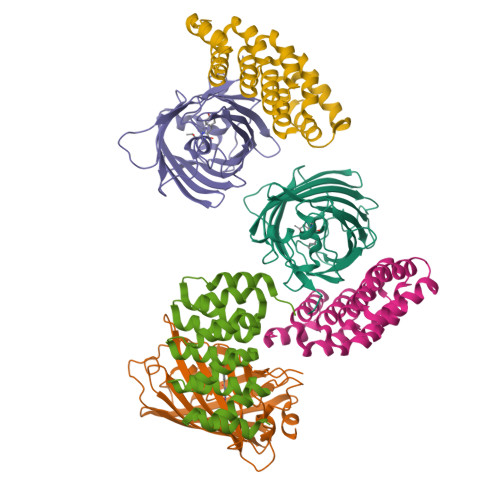
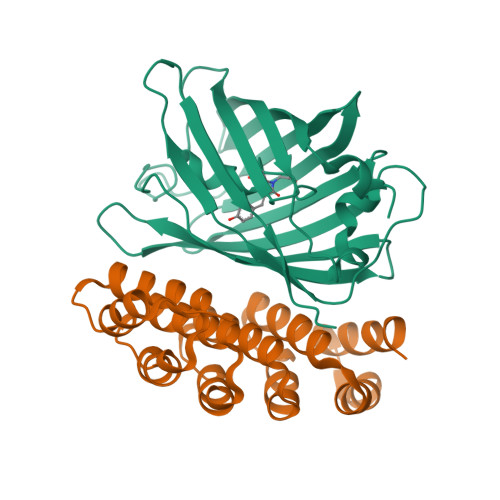
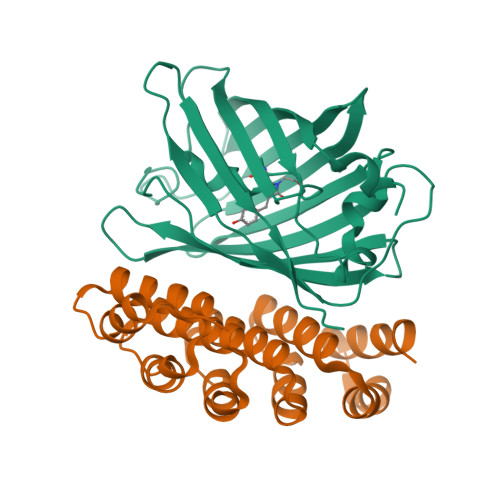
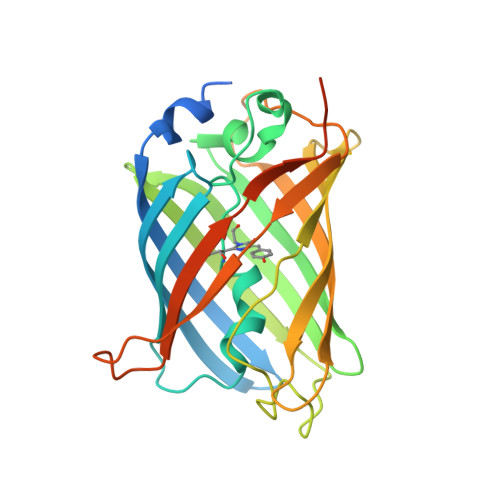
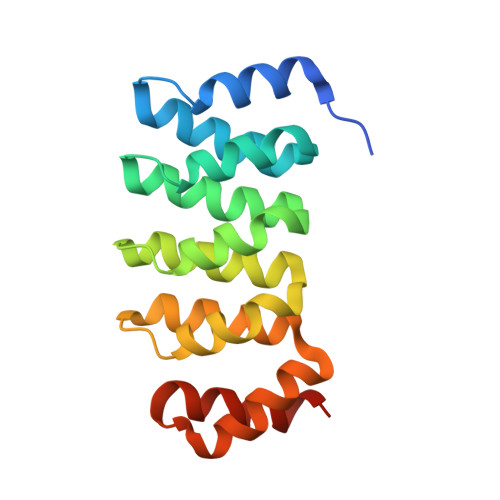
A family of artificial proteins, named αRep, based on a natural family of helical repeat was previously designed. αRep members are efficiently expressed, folded and extremely stable proteins. A large αRep library was constructed creating proteins with a randomized interaction surface. In the present study, we show that the αRep library is an efficient source of tailor-made specific proteins with direct applications in biochemistry and cell biology. From this library, we selected by phage display αRep binders with nanomolar dissociation constants against the GFP. The structures of two independent αRep binders in complex with the GFP target were solved by X-ray crystallography revealing two totally different binding modes. The affinity of the selected αReps for GFP proved sufficient for practically useful applications such as pull-down experiments. αReps are disulfide free proteins and are efficiently and functionally expressed in eukaryotic cells: GFP-specific αReps are clearly sequestrated by their cognate target protein addressed to various cell compartments. These results suggest that αRep proteins with tailor-made specificity can be selected and used in living cells to track, modulate or interfere with intracellular processes.
Organizational Affiliation:
Institute for Integrative Biology of the Cell (I2BC), UMR 9198, CEA, CNRS, Université Paris-Sud, Bât 430, 91405 Orsay Cedex, France.