Crystal Structure of Human Myotubularin-Related Protein 1 Provides Insight into the Structural Basis of Substrate Specificity
Bong, S.M., Son, K.B., Yang, S.W., Park, J.W., Cho, J.W., Kim, K.T., Kim, H., Kim, S.J., Kim, Y.J., Lee, B.I.(2016) PLoS One 11: e0152611-e0152611
- PubMed: 27018598
- DOI: https://doi.org/10.1371/journal.pone.0152611
- Primary Citation of Related Structures:
5C16 - PubMed Abstract:
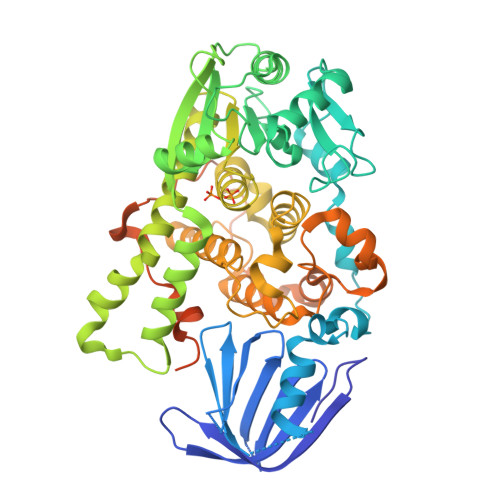
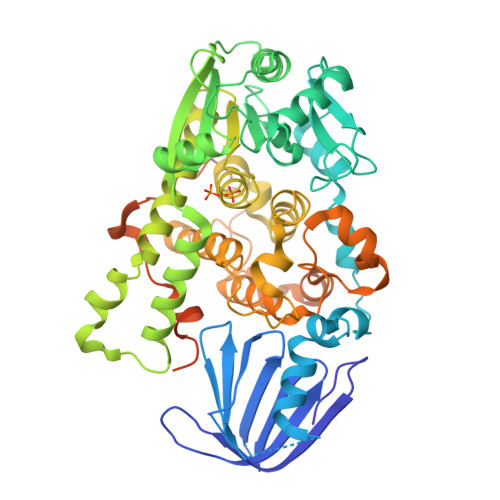
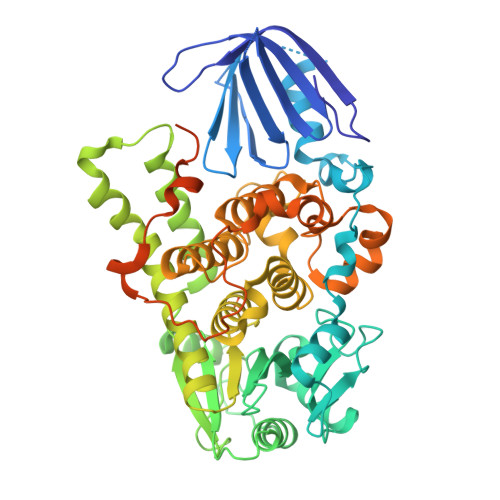
Myotubularin-related protein 1 (MTMR1) is a phosphatase that belongs to the tyrosine/dual-specificity phosphatase superfamily. MTMR1 has been shown to use phosphatidylinositol 3-monophosphate (PI(3)P) and/or phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) as substrates. Here, we determined the crystal structure of human MTMR1. The refined model consists of the Pleckstrin homology (PH)-GRAM and phosphatase (PTP) domains. The overall structure was highly similar to the previously reported MTMR2 structure. Interestingly, two phosphate molecules were coordinated by strictly conserved residues located in the C(X)5R motif of the active site. Additionally, our biochemical studies confirmed the substrate specificity of MTMR1 for PI(3)P and PI(3,5)P2 over other phosphatidylinositol phosphates. Our structural and enzymatic analyses provide insight into the catalytic mechanism and biochemical properties of MTMR1.
Organizational Affiliation:
Research institute, National Cancer Center, Goyang, Gyeonggi 10408, Republic of Korea.