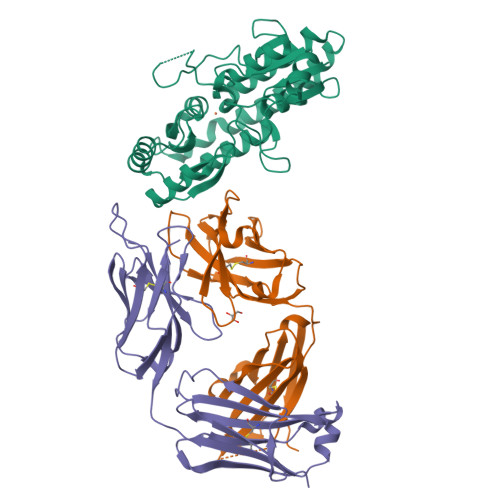
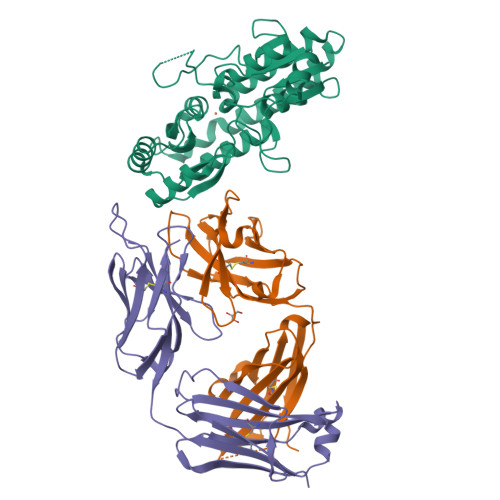
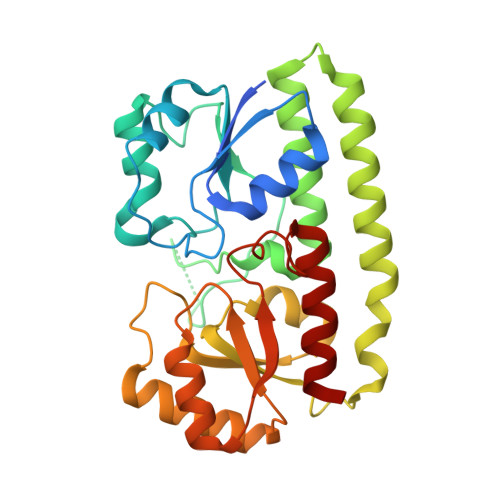
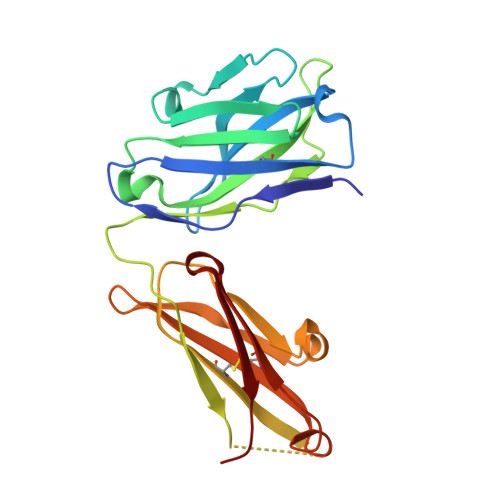
High Resolution Mapping of Bactericidal Monoclonal Antibody Binding Epitopes on Staphylococcus aureus Antigen MntC.
Gribenko, A.V., Parris, K., Mosyak, L., Li, S., Handke, L., Hawkins, J.C., Severina, E., Matsuka, Y.V., Anderson, A.S.(2016) PLoS Pathog 12: e1005908-e1005908
- PubMed: 27689696
- DOI: https://doi.org/10.1371/journal.ppat.1005908
- Primary Citation of Related Structures:
5HDQ - PubMed Abstract:
The Staphylococcus aureus manganese transporter protein MntC is under investigation as a component of a prophylactic S.aureus vaccine. Passive immunization with monoclonal antibodies mAB 305-78-7 and mAB 305-101-8 produced using MntC was shown to significantly reduce S. aureus burden in an infant rat model of infection. Earlier interference mapping suggested that a total of 23 monoclonal antibodies generated against MntC could be subdivided into three interference groups, representing three independent immunogenic regions. In the current work binding epitopes for selected representatives of each of these interference groups (mAB 305-72-5 - group 1, mAB 305-78-7 - group 2, and mAB 305-101-8 - group 3) were mapped using Hydrogen-Deuterium Exchange Mass Spectrometry (DXMS). All of the identified epitopes are discontinuous, with binding surface formed by structural elements that are separated within the primary sequence of the protein but adjacent in the context of the three-dimensional structure. The approach was validated by co-crystallizing the Fab fragment of one of the antibodies (mAB 305-78-7) with MntC and solving the three-dimensional structure of the complex. X-ray results themselves and localization of the mAB 305-78-7 epitope were further validated using antibody binding experiments with MntC variants containing substitutions of key amino acid residues. These results provided insight into the antigenic properties of MntC and how these properties may play a role in protecting the hostagainst S. aureus infection by preventing the capture and transport of Mn2+, a key element that the pathogen uses to evade host immunity.
Organizational Affiliation:
Pfizer Vaccine Research and Development, Pearl River, New York, United States of America.