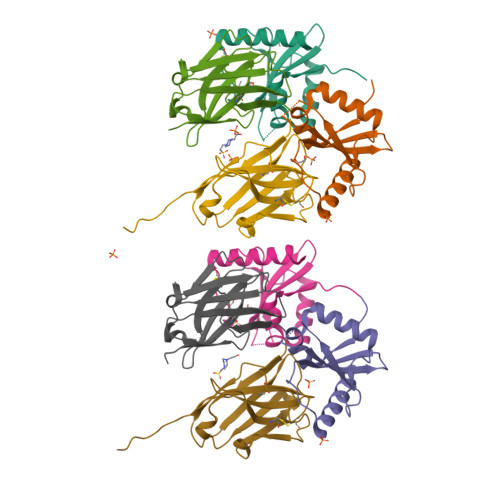
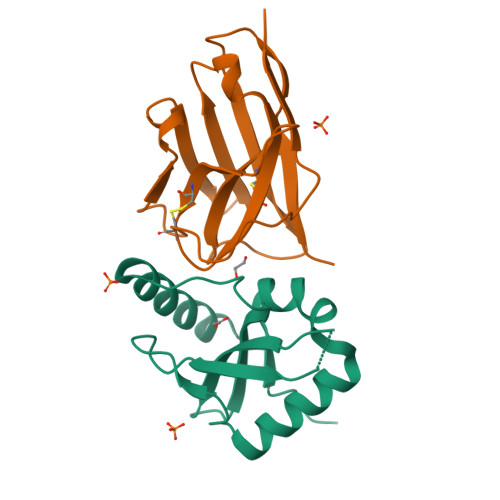
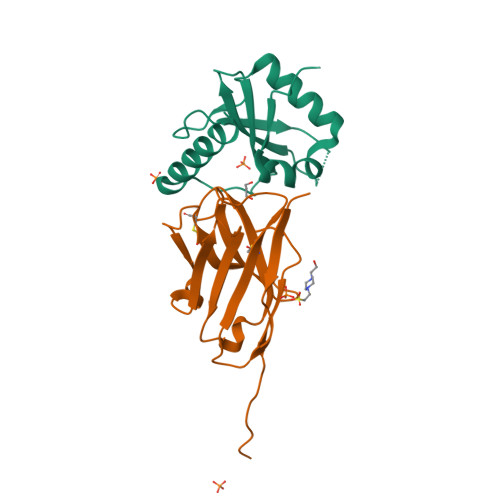
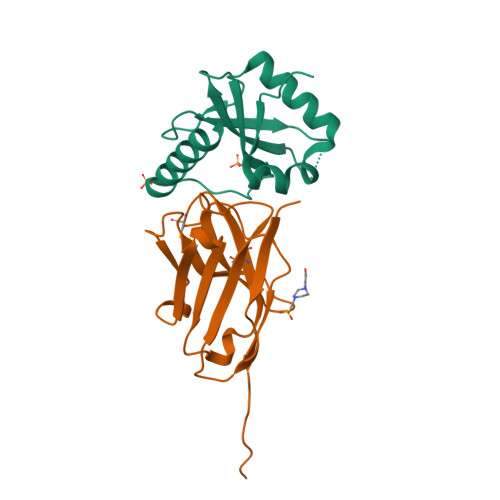
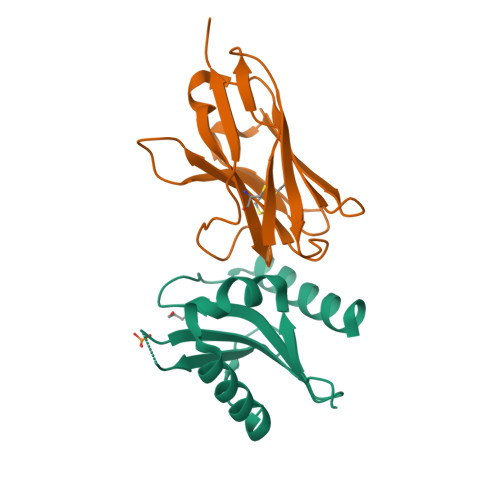
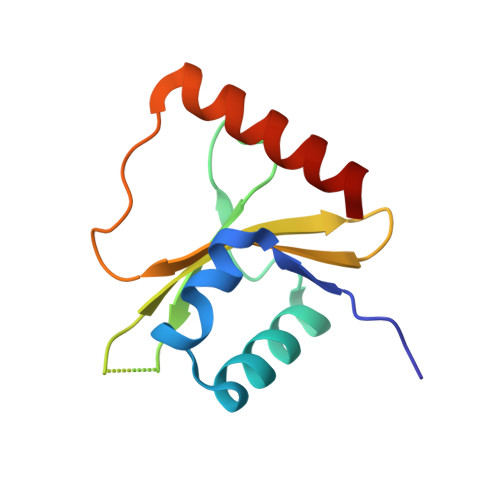
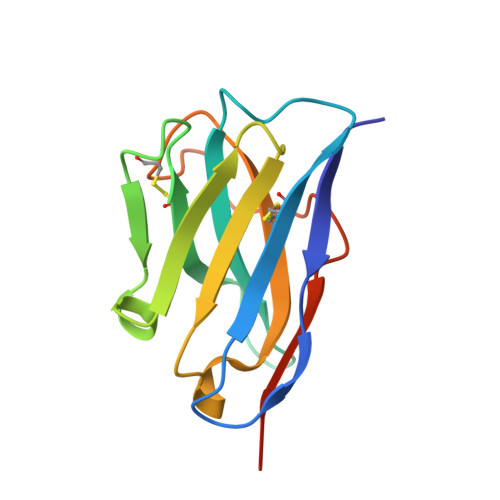
Ribosome-dependent Vibrio cholerae mRNAse HigB2 is regulated by a beta-strand sliding mechanism.
Hadzi, S., Garcia-Pino, A., Haesaerts, S., Jurenas, D., Gerdes, K., Lah, J., Loris, R.(2017) Nucleic Acids Res 45: 4972-4983
- PubMed: 28334932
- DOI: https://doi.org/10.1093/nar/gkx138
- Primary Citation of Related Structures:
5J9I, 5JA8, 5JA9, 5JAA, 5MJE - PubMed Abstract:
Toxin-antitoxin (TA) modules are small operons involved in bacterial stress response and persistence. higBA operons form a family of TA modules with an inverted gene organization and a toxin belonging to the RelE/ParE superfamily. Here, we present the crystal structures of chromosomally encoded Vibrio cholerae antitoxin (VcHigA2), toxin (VcHigB2) and their complex, which show significant differences in structure and mechanisms of function compared to the higBA module from plasmid Rts1, the defining member of the family. The VcHigB2 is more closely related to Escherichia coli RelE both in terms of overall structure and the organization of its active site. VcHigB2 is neutralized by VcHigA2, a modular protein with an N-terminal intrinsically disordered toxin-neutralizing segment followed by a C-terminal helix-turn-helix dimerization and DNA binding domain. VcHigA2 binds VcHigB2 with picomolar affinity, which is mainly a consequence of entropically favorable de-solvation of a large hydrophobic binding interface and enthalpically favorable folding of the N-terminal domain into an α-helix followed by a β-strand. This interaction displaces helix α3 of VcHigB2 and at the same time induces a one-residue shift in the register of β-strand β3, thereby flipping the catalytically important Arg64 out of the active site.
Organizational Affiliation:
Structural Biology Brussels, Department of Biotechnology, Vrije Universiteit Brussel, B-1050 Brussel, Belgium.