Cergutuzumab amunaleukin (CEA-IL2v), a CEA-targeted IL-2 variant-based immunocytokine for combination cancer immunotherapy: Overcoming limitations of aldesleukin and conventional IL-2-based immunocytokines.
Klein, C., Waldhauer, I., Nicolini, V.G., Freimoser-Grundschober, A., Nayak, T., Vugts, D.J., Dunn, C., Bolijn, M., Benz, J., Stihle, M., Lang, S., Roemmele, M., Hofer, T., van Puijenbroek, E., Wittig, D., Moser, S., Ast, O., Brunker, P., Gorr, I.H., Neumann, S., de Vera Mudry, M.C., Hinton, H., Crameri, F., Saro, J., Evers, S., Gerdes, C., Bacac, M., van Dongen, G., Moessner, E., Umana, P.(2017) Oncoimmunology 6: e1277306-e1277306
- PubMed: 28405498
- DOI: https://doi.org/10.1080/2162402X.2016.1277306
- Primary Citation of Related Structures:
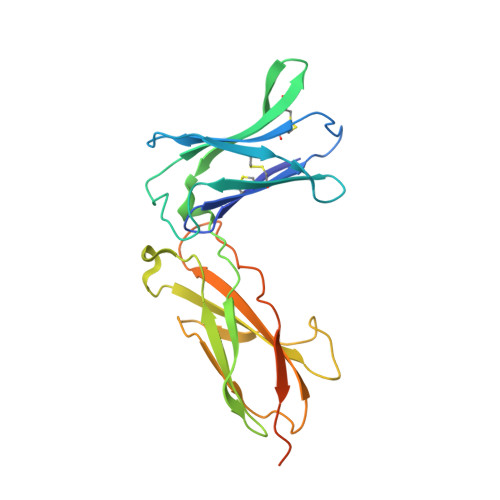
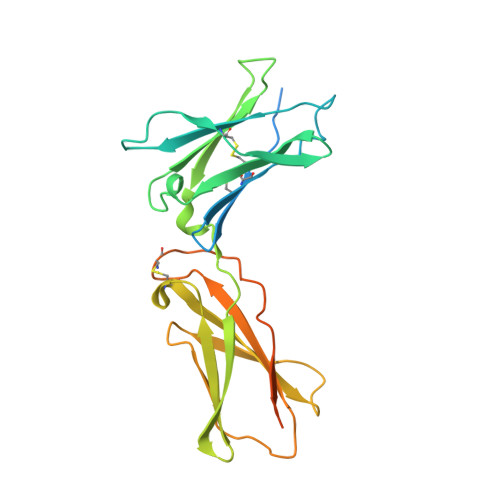
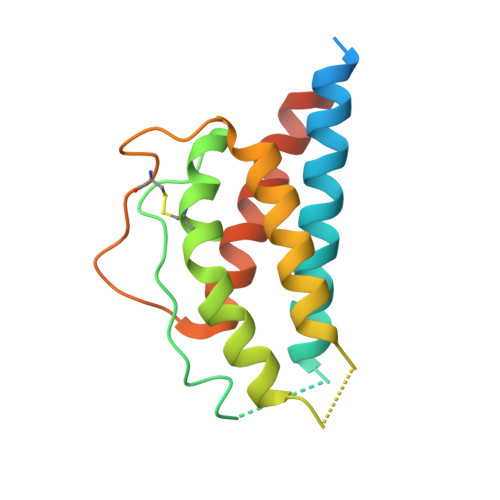
5M5E - PubMed Abstract:
We developed cergutuzumab amunaleukin (CEA-IL2v, RG7813), a novel monomeric CEA-targeted immunocytokine, that comprises a single IL-2 variant (IL2v) moiety with abolished CD25 binding, fused to the C-terminus of a high affinity, bivalent carcinoembryonic antigen (CEA)-specific antibody devoid of Fc-mediated effector functions. Its molecular design aims to (i) avoid preferential activation of regulatory T-cells vs. immune effector cells by removing CD25 binding; (ii) increase the therapeutic index of IL-2 therapy by (a) preferential retention at the tumor by having a lower dissociation rate from CEA-expressing cancer cells vs. IL-2R-expressing cells, (b) avoiding any FcγR-binding and Fc effector functions and (c) reduced binding to endothelial cells expressing CD25; and (iii) improve the pharmacokinetics, and thus convenience of administration, of IL-2. The crystal structure of the IL2v-IL-2Rβγ complex was determined and CEA-IL2v activity was assessed using human immune effector cells. Tumor targeting was investigated in tumor-bearing mice using 89 Zr-labeled CEA-IL2v. Efficacy studies were performed in (a) syngeneic mouse models as monotherapy and combined with anti-PD-L1, and in (b) xenograft mouse models in combination with ADCC-mediating antibodies. CEA-IL2v binds to CEA with pM avidity but not to CD25, and consequently did not preferentially activate Tregs. In vivo , CEA-IL2v demonstrated superior pharmacokinetics and tumor targeting compared with a wild-type IL-2-based CEA immunocytokine (CEA-IL2wt). CEA-IL2v strongly expanded NK and CD8 + T cells, skewing the CD8 + :CD4 + ratio toward CD8 + T cells both in the periphery and in the tumor, and mediated single agent efficacy in syngeneic MC38-CEA and PancO2-CEA models. Combination with trastuzumab, cetuximab and imgatuzumab, all of human IgG1 isotype, resulted in superior efficacy compared with the monotherapies alone. Combined with anti-PD-L1, CEA-IL2v mediated superior efficacy over the respective monotherapies, and over the combination with an untargeted control immunocytokine. These preclinical data support the ongoing clinical investigation of the cergutuzumab amunaleukin immunocytokine with abolished CD25 binding for the treatment of CEA-positive solid tumors in combination with PD-L1 checkpoint blockade and ADCC competent antibodies.
Organizational Affiliation:
Roche Pharma Research & Early Development, Roche Innovation Center Zurich , Schlieren, Switzerland.