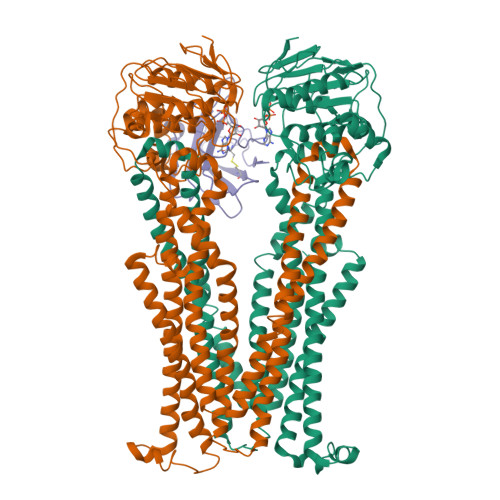
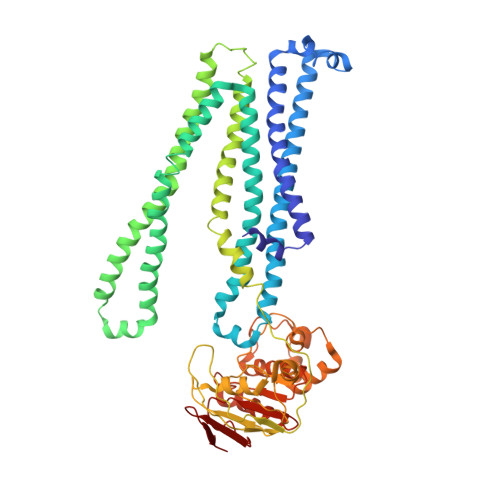
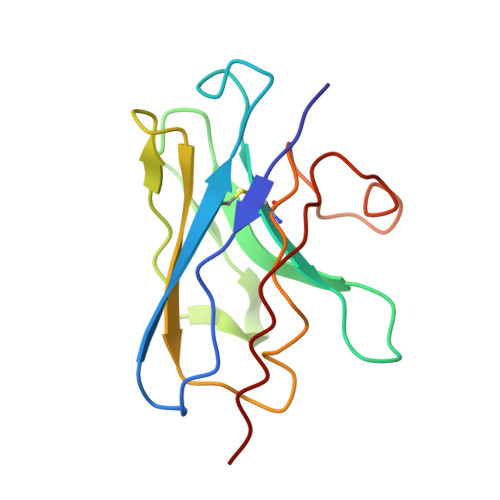
Structural basis of inhibition of lipid-linked oligosaccharide flippase PglK by a conformational nanobody.
Perez, C., Kohler, M., Janser, D., Pardon, E., Steyaert, J., Zenobi, R., Locher, K.P.(2017) Sci Rep 7: 46641-46641
- PubMed: 28422165
- DOI: https://doi.org/10.1038/srep46641
- Primary Citation of Related Structures:
5NBD - PubMed Abstract:
PglK is an ABC transporter that flips a lipid-linked oligosaccharide (LLO) that serves as a donor in protein N-glycosylation. Previous structures revealed two inward-facing conformations, both with very large separations of the nucleotide binding domains (NBDs), and a closed, ADP-bound state that featured an occluded cavity. To investigate additional states, we developed conformation-sensitive, single-domain camelid nanobodies (Nb) and studied their effect on PglK activity. Biochemical, structural, and mass spectrometric analyses revealed that one inhibitory Nb binds as a single copy to homodimeric PglK. The co-crystal structure of this Nb and ADP-bound PglK revealed a new, narrowly inward-open conformation. Rather than inducing asymmetry in the PglK homodimer, the binding of one Nb results in steric constraints that prevent a second Nb to access the symmetry-related site in PglK. The Nb performed its inhibitory role by a "sticky-doorstop" mechanism, where inhibition of ATP hydrolysis and LLO flipping activity occurs due to impaired closing of the NBD interface, which prevents PglK from converting to an outward-open conformation. This inhibitory mode suggests tight conformational coupling between the ATPase sites, which may apply to other ABC transporters.
Organizational Affiliation:
Department of Biology, Institute of Molecular Biology and Biophysics, ETH Zürich, CH-8093 Zürich, Switzerland.