Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors.
Zhou, X.E., He, Y., de Waal, P.W., Gao, X., Kang, Y., Van Eps, N., Yin, Y., Pal, K., Goswami, D., White, T.A., Barty, A., Latorraca, N.R., Chapman, H.N., Hubbell, W.L., Dror, R.O., Stevens, R.C., Cherezov, V., Gurevich, V.V., Griffin, P.R., Ernst, O.P., Melcher, K., Xu, H.E.(2017) Cell 170: 457-469.e13
- PubMed: 28753425
- DOI: https://doi.org/10.1016/j.cell.2017.07.002
- Primary Citation of Related Structures:
5W0P - PubMed Abstract:
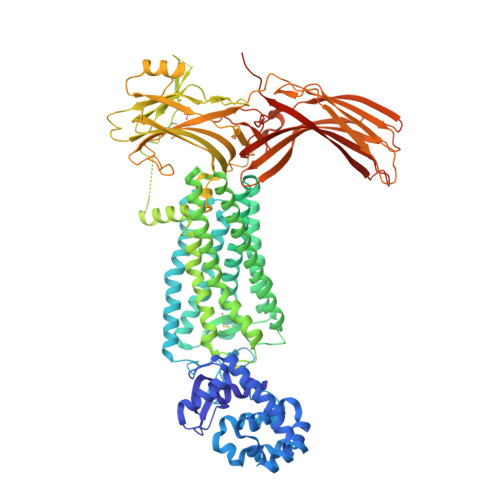
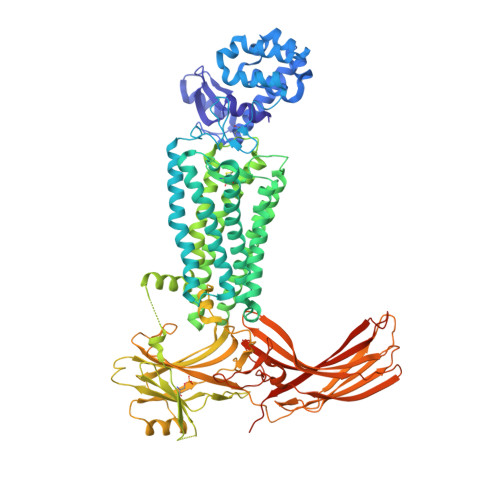
G protein-coupled receptors (GPCRs) mediate diverse signaling in part through interaction with arrestins, whose binding promotes receptor internalization and signaling through G protein-independent pathways. High-affinity arrestin binding requires receptor phosphorylation, often at the receptor's C-terminal tail. Here, we report an X-ray free electron laser (XFEL) crystal structure of the rhodopsin-arrestin complex, in which the phosphorylated C terminus of rhodopsin forms an extended intermolecular β sheet with the N-terminal β strands of arrestin. Phosphorylation was detected at rhodopsin C-terminal tail residues T336 and S338. These two phospho-residues, together with E341, form an extensive network of electrostatic interactions with three positively charged pockets in arrestin in a mode that resembles binding of the phosphorylated vasopressin-2 receptor tail to β-arrestin-1. Based on these observations, we derived and validated a set of phosphorylation codes that serve as a common mechanism for phosphorylation-dependent recruitment of arrestins by GPCRs.
Organizational Affiliation:
VARI-SIMM Center, Center for Structure and Function of Drug Targets, CAS-Key Laboratory of Receptor Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China; Laboratory of Structural Sciences, Center for Structural Biology and Drug Discovery, Van Andel Research Institute, Grand Rapids, MI 49503, USA.