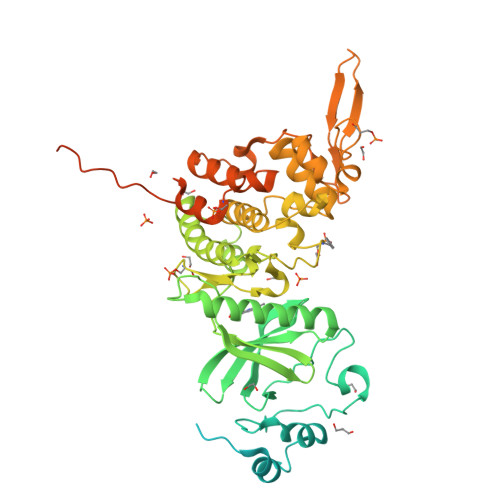
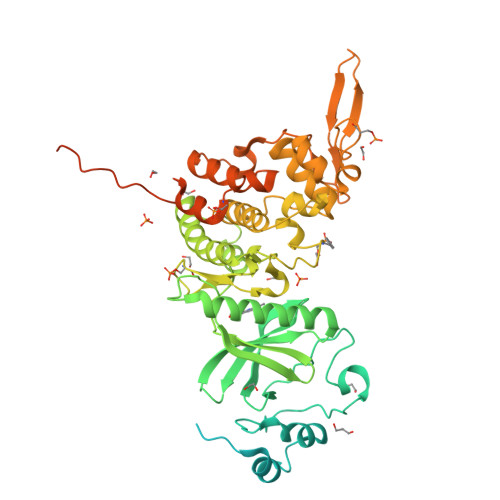
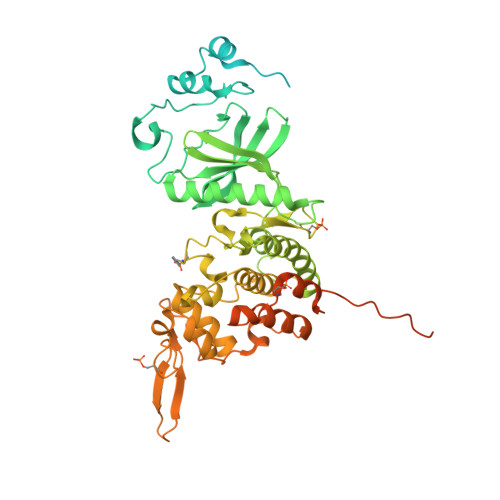
Crystal Structure of Human Dual-Specificity Tyrosine-Regulated Kinase 3 Reveals New Structural Features and Insights into its Auto-phosphorylation
Kim, K., Cha, J.S., Cho, Y.S., Kim, H., Chang, N., Kim, H.J., Cho, H.S.(2018) J Mol Biology 430: 1521-1530
- PubMed: 29634919
- DOI: https://doi.org/10.1016/j.jmb.2018.04.001
- Primary Citation of Related Structures:
5Y86 - PubMed Abstract:
Dual-specificity tyrosine-regulated kinases (DYRKs) auto-phosphorylate a critical tyrosine residue in their activation loop and phosphorylate their substrate on serine and threonine residues. The auto-phosphorylation occurs intramolecularly and is a one-off event. DYRK3 is selectively expressed at a high level in hematopoietic cells and attenuates erythroblast development, leading to anemia. In the present study, we determined the crystal structure of the mature form of human DYRK3 in complex with harmine, an ATP competitive inhibitor. The crystal structure revealed a phosphorylation site, residue S350, whose phosphorylation increases the stability of DYRK3 and enhances its kinase activity. In addition, our structural and biochemical assays suggest that the N-terminal auto-phosphorylation accessory domain stabilizes the DYRK3 protein, followed by auto-phosphorylation of the tyrosine of the activation loop, which is important for kinase activity. Finally, our docking analysis provides information for the design of novel and potent therapeutics to treat anemia.
Organizational Affiliation:
Department of Systems Biology and Division of Life Sciences, Yonsei University, 50 Yonsei-ro, Seodaemun-gu, Seoul 03722, Republic of Korea.