Design, Synthesis, and Evaluation of Piperazinyl Pyrrolidin-2-ones as a Novel Series of Reversible Monoacylglycerol Lipase Inhibitors
Aida, J., Fushimi, M., Kusumoto, T., Sugiyama, H., Arimura, N., Ikeda, S., Sasaki, M., Sogabe, S., Aoyama, K., Koike, T.(2018) J Med Chem 61: 9205-9217
- PubMed: 30251836
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00824
- Primary Citation of Related Structures:
5ZUN - PubMed Abstract:
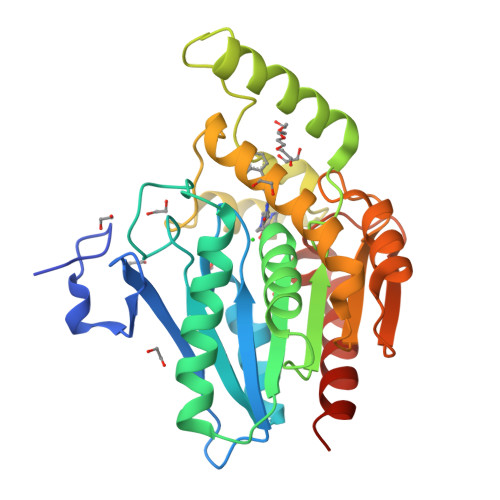
Monoacylglycerol lipase (MAGL) is a major serine hydrolase that hydrolyzes 2-arachidonoylglycerol (2-AG) to arachidonic acid (AA) and glycerol in the brain. Because 2-AG and AA are endogenous biologically active ligands in the brain, inhibition of MAGL is an attractive therapeutic target for CNS disorders, particularly neurodegenerative diseases. In this study, we report the structure-based drug design of novel piperazinyl pyrrolidin-2-ones starting from our hit compounds 2a and 2b. By enhancing the interaction of the piperazinyl pyrrolidin-2-one core and its substituents with the MAGL enzyme via design modifications, we identified a potent and reversible MAGL inhibitor, compound ( R)-3t. Oral administration of compound ( R)-3t to mice decreased AA levels and elevated 2-AG levels in the brain.
Organizational Affiliation:
Research , Takeda Pharmaceutical Co., Ltd. , 26-1, Muraoka-Higashi 2-Chome , Fujisawa , Kanagawa 251-8555 , Japan.