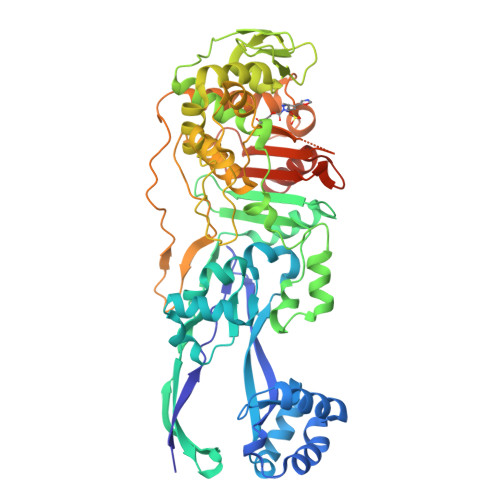
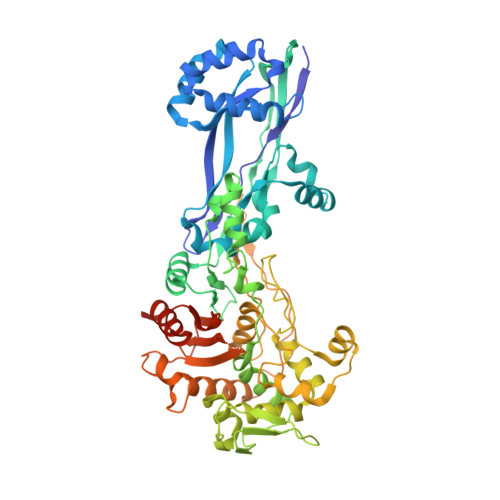
Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
Levy, N., Bruneau, J.M., Le Rouzic, E., Bonnard, D., Le Strat, F., Caravano, A., Chevreuil, F., Barbion, J., Chasset, S., Ledoussal, B., Moreau, F., Ruff, M.(2019) J Med Chem 62: 4742-4754
- PubMed: 30995398
- DOI: https://doi.org/10.1021/acs.jmedchem.9b00338
- Primary Citation of Related Structures:
6G9F, 6G9P, 6G9S - PubMed Abstract:
Penicillin-binding proteins (PBPs) are the targets of the β-lactams, the most successful class of antibiotics ever developed against bacterial infections. Unfortunately, the worldwide and rapid spread of large spectrum β-lactam resistance genes such as carbapenemases is detrimental to the use of antibiotics in this class. New potent PBP inhibitors are needed, especially compounds that resist β-lactamase hydrolysis. Here we describe the structure of the E. coli PBP2 in its Apo form and upon its reaction with 2 diazabicyclo derivatives, avibactam and CPD4, a new potent PBP2 inhibitor. Examination of these structures shows that unlike avibactam, CPD4 can perform a hydrophobic stacking on Trp370 in the active site of E. coli PBP2. This result, together with sequence analysis, homology modeling, and SAR, allows us to propose CPD4 as potential starting scaffold to develop molecules active against a broad range of bacterial species at the top of the WHO priority list.
Organizational Affiliation:
Mutabilis , 102 Avenue Gaston Roussel , 93230 Romainville , France.