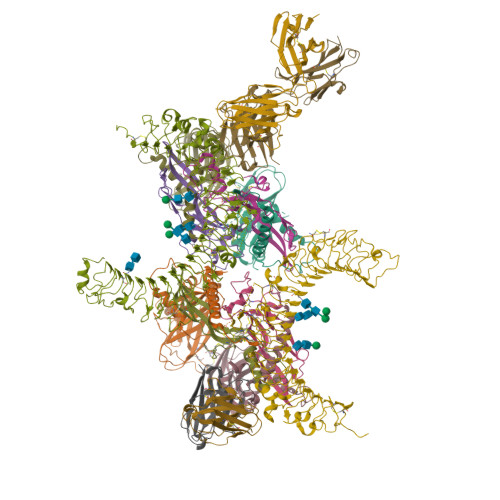
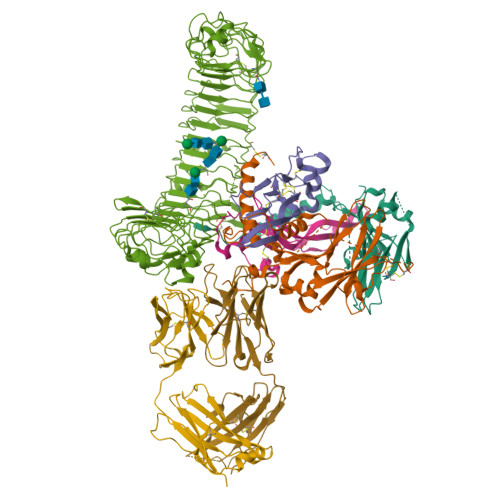
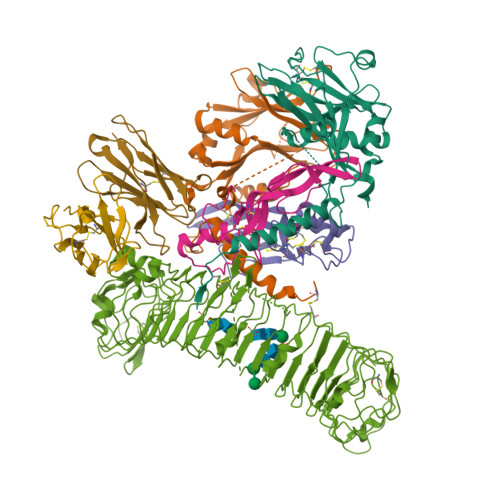
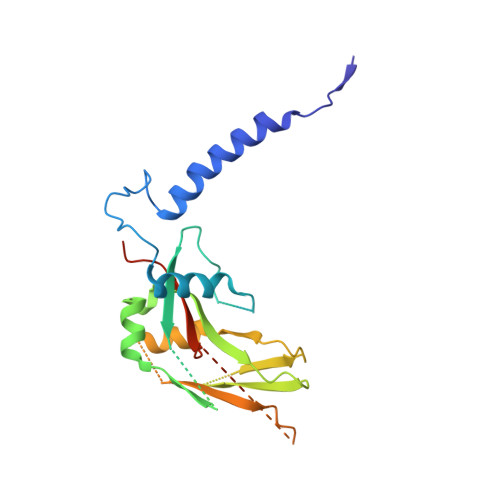
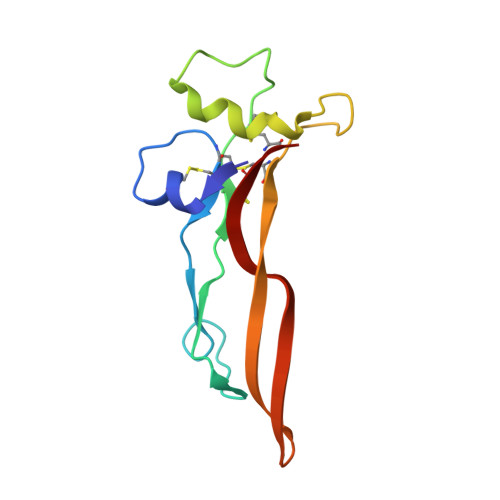
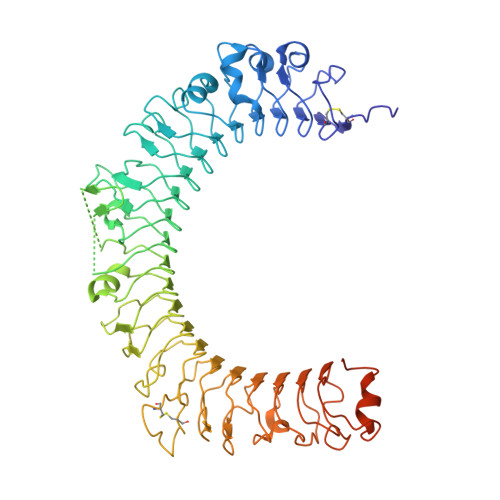
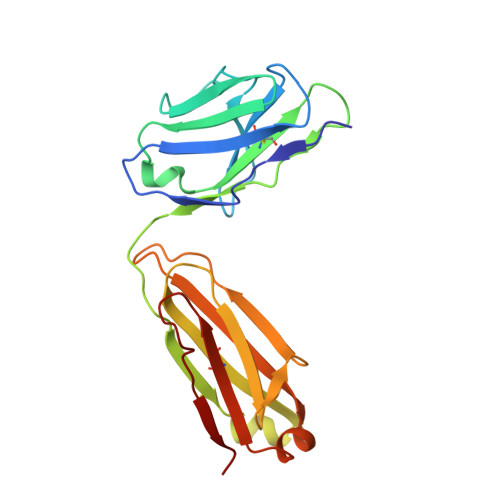
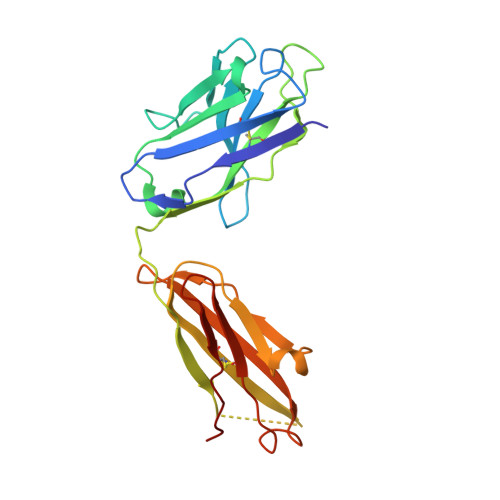
Structural basis of latent TGF-beta 1 presentation and activation by GARP on human regulatory T cells.
Lienart, S., Merceron, R., Vanderaa, C., Lambert, F., Colau, D., Stockis, J., van der Woning, B., De Haard, H., Saunders, M., Coulie, P.G., Savvides, S.N., Lucas, S.(2018) Science 362: 952-956
- PubMed: 30361387
- DOI: https://doi.org/10.1126/science.aau2909
- Primary Citation of Related Structures:
6GFF - PubMed Abstract:
Transforming growth factor-β1 (TGF-β1) is one of very few cytokines produced in a latent form, requiring activation to exert any of its vastly diverse effects on development, immunity, and cancer. Regulatory T cells (T regs ) suppress immune cells within close proximity by activating latent TGF-β1 presented by GARP (glycoprotein A repetitions predominant) to integrin αVβ8 on their surface. We solved the crystal structure of GARP:latent TGF-β1 bound to an antibody that stabilizes the complex and blocks release of active TGF-β1. This finding reveals how GARP exploits an unusual medley of interactions, including fold complementation by the amino terminus of TGF-β1, to chaperone and orient the cytokine for binding and activation by αVβ8. Thus, this work further elucidates the mechanism of antibody-mediated blockade of TGF-β1 activation and immunosuppression by T regs .
Organizational Affiliation:
de Duve Institute, UCLouvain, 1200 Brussels, Belgium.